We had a paper in Science go online on May 25, 2021: Estimating infectiousness throughout SARS-CoV-2 infection course.

Pages
Archives
- July 2021
- April 2021
- October 2019
- July 2018
- August 2017
- July 2017
- September 2016
- June 2016
- December 2014
- January 2014
- December 2013
- September 2013
- July 2013
- June 2013
- May 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- August 2012
- May 2012
- April 2012
- February 2012
- January 2012
- November 2011
- October 2011
- August 2011
- July 2011
- June 2011
- March 2011
- February 2011
- January 2011
- November 2010
- September 2010
- August 2010
- July 2010
- June 2010
- January 2010
- November 2009
- October 2009
- September 2009
- August 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- June 2007
- May 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
Categories
Meta
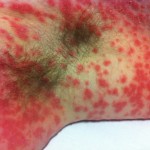
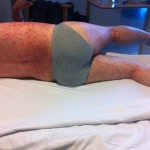
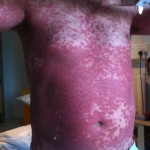
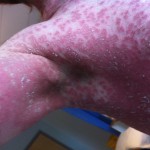
Estimating infectiousness throughout SARS-CoV-2 infection course
23:13 July 16th, 2021 by terry. Posted under me. | Comments Off on Estimating infectiousness throughout SARS-CoV-2 infection course
The vikings had smallpox
08:58 April 18th, 2021 by terry. Posted under me. | Comments Off on The vikings had smallpox
In July 2020 we published a paper in Science on ancient smallpox viruses: Diverse variola virus (smallpox) strains were widespread in northern Europe in the Viking Age. That was a couple of years of work.
There were some write-ups, e.g. Viking Age Smallpox Complicates Story of Viral Evolution.
The paper was written in collaboration with Eske Willerslev and his GeoGenetics group in Copenhagen.
Daudin – a Python shell
23:11 October 13th, 2019 by terry. Posted under me, programming, python, tech. | 1 Comment »
A few nights ago I wrote daudin, a command-line shell based on Python. It allows you to easily mix UNIX and Python on the command line.
Source code and documentation: https://github.com/terrycojones/daudin.
Install via pip install daudin.
Daudin was a French zoologist who named the Python genus in 1826.
Papers on ancient hepatitis B virus and human parvovirus B19
17:56 July 15th, 2018 by terry. Posted under me. | Comments Off on Papers on ancient hepatitis B virus and human parvovirus B19
We just published two papers.
Ancient hepatitis B viruses from the Bronze Age to the Medieval period in Nature, and Ancient human parvovirus B19 in Eurasia reveals its long-term association with humans in PNAS.
These were written in collaboration with Eske Willerslev and his GeoGenetics group in Copenhagen.
Barbara Mühlemann and I were on the BBC Breakfast show the morning after the HBV paper came out, if you feel like watching.
A BLAST puzzle
22:25 August 25th, 2017 by terry. Posted under me. | Comments Off on A BLAST puzzle
Re-posted from my internal Cambridge notebook.
I have a strange situation. I have two almost identical BLAST databases (20170821 and 20170824). They both contain a certain subject sequence LC074724.
I query them both, using BLAST’s default search, with an identical query, Q. In one case (20170824) there is a very strong match but in the other (20170821) the query is not matched at all.
I’m trying to figure out why.
BLAST version
$ blastn -version
blastn: 2.6.0+
Package: blast 2.6.0, build May 11 2017 22:22:40
The subject sequence is the same in both databases
Both databases contain an identical subject sequence:
$ blastdbcmd -entry LC074724 -db 20170824 | md5
fcfbc167c7f81dfd75aad1bf3db6220b
$ blastdbcmd -entry LC074724 -db 20170821 | md5
fcfbc167c7f81dfd75aad1bf3db6220b
Doing a match
There is a good match of Q against LC074724 in 20170824:
$ blastn -outfmt '6 qaccver saccver bitscore' -db 20170824 -query Q.fasta \
-task blastn -max_hsps 1 | grep LC074724
Q LC074724.1 2581
Note that the bitscore of the LC074724 match is 2581.
But there is no match of LC074724 at all in 20170821:
$ blastn -outfmt '6 qaccver saccver bitscore' -db 20170821 -query Q.fasta \
-task blastn -max_hsps 1 | grep LC074724
The option -outfmt '6 qaccver saccver bitscore' tells BLAST to print the query accession number and version, the subject accession and version, and the bitscore of the match.
What are the best hits for 20170821
So what are the bitscores of the Q matches in 20170821?
$ blastn -outfmt '6 bitscore' -db 20170821 -query Q.fasta -task blastn -max_hsps 1 | sort -nr | head
3301
2616
2590
2590
2563
2554
2547
2544
2540
2540
The bitscore of Q against LC074724 in 20170824 (2581) would have placed it in 5th position in the top 10 matches of Q in 20170821, but LC074724 is not matched at all in that database! The worst reported match in 20170821 has bitscore 2354.
What’s the difference between the databases?
Get the ids of the subjects in each database
$ blastdbcmd -entry all -db 20170824 | egrep '^>' | cut -c2- | sort > 20170824.ids
$ blastdbcmd -entry all -db 20170821 | egrep '^>' | cut -c2- | sort > 20170821.ids
$ wc -l 2017082?.ids
9754 20170821.ids
9111 20170824.ids
18865 total
20170821 has 643 sequences that are not in 20170824:
$ comm -23 20170821.ids 20170824.ids | wc -l
643
20170824 has no sequences that are not in 20170821:
$ comm -13 20170821.ids 20170824.ids | wc -l
0
and the two have 9111 sequences in common:
$ comm -12 20170821.ids 20170824.ids | wc -l
9111
These numbers match with the numbers from wc -l above.
So 20170821 is just 20170824 with an extra 643 sequences.
In other words, just adding some sequences to 20170824 to produce 20170821 makes one of the top hits completely vanish from the output!
Maybe LC074724 matches, but with a low bit score?
BLAST only shows the top 500 matched subjects by default. Let’s ask it to show more:
$ blastn -outfmt '6 qaccver saccver bitscore' -db 20170821 -query Q.fasta \
-task blastn -max_hsps 1 -max_target_seqs 10000 > 20170821-10000-matches.txt
And….
$ head 20170821-10000-matches.txt
Q Q 3301
Q X234A 2616
Q CS388973.1 2590
Q DM059402.1 2590
Q LC074724.1 2581
Q KJ843188.1 2576
Q AB697496.1 2576
Q AB697499.1 2576
Q AB697503.1 2576
Q AB697508.1 2576
Holy shit, there it is! And it’s in 5th position, just as it should be (based on the bitscores discussed above), and with exactly the same high score that it receives when matched in the other database (20170824).
Just to confirm, taking away the -max_target_seqs option gets no match:
$ blastn -outfmt '6 qaccver saccver bitscore' -db 20170821 -query Q.fasta \
-task blastn -max_hsps 1 | grep LC074724
$
HOW ON EARTH CAN THAT BE HAPPENING?
Three confusingly-named BLAST options
I’ve always found the names and descriptions of the following three BLAST command-line options very confusing.
From the Formatting options section of blastn -help:
-num_descriptions <Integer, >=0>
Number of database sequences to show one-line descriptions for
Not applicable for outfmt > 4
Default = `500'
* Incompatible with: max_target_seqs
-num_alignments <Integer, >=0>
Number of database sequences to show alignments for
Default = `250'
* Incompatible with: max_target_seqs
And from the Restrict search or results section:
-max_target_seqs <Integer, >=1>
Maximum number of aligned sequences to keep
Not applicable for outfmt <= 4
Default = `500'
* Incompatible with: num_descriptions, num_alignments
From the section names, it seems like only the latter option (-max_target_seqs) would have any effect on the search, and that the former two are just about what is displayed.
But… using either just -num_descriptions or -num_alignments with a big value also gets a match:
$ blastn -db 20170821 -query Q.fasta -task blastn -max_hsps 1 -num_descriptions 10000 | grep LC074724
LC074724.1 Hepatitis B virus DNA, complete genome, isolate: p621 2581 0.0
>LC074724.1 Hepatitis B virus DNA, complete genome, isolate: p621
$ blastn -db 20170821 -query Q.fasta -task blastn -max_hsps 1 -num_alignments 10000 | grep LC074724
LC074724.1 Hepatitis B virus DNA, complete genome, isolate: p621 2581 0.0
>LC074724.1 Hepatitis B virus DNA, complete genome, isolate: p621
So BLAST is apparently finding the LC074724 match even without the high -max_target_seqs option, but it’s not displaying it unless there is a high -num_alignments or -num_descriptions value.
That makes no sense at all. BLAST is finding the match without the -max_target_seqs option and the match has the 5th highest score, so why is it not being displayed?
I don’t understand the difference between -num_alignments and -num_descriptions.
Maybe -num_alignments is per query and -num_descriptions is in the overall result set (i.e., across all queries). BLAST seems to use “alignment” to refer to a match between a query and a single subject (and that match may have multiple high-scoring pairs (HSPs), each with its own bit score). But why is there a -num_descriptions option? I can only think that BLAST is allocating memory to hold subject descriptions and this option is used to allocate some fixed storage for the overall search. So, possibly, if the -num_descriptions limit is reached during the search, BLAST gives up because it knows it cannot store more subject descriptions. But only setting -num_alignments also gets me a match, so maybe BLAST silently raises num_descriptions to be at least as big as num_alignments. I don’t know.
Just to confirm, when none of the 3 options are given and no -outfmt
option is either, the match is not found:
$ blastn -db 20170821 -query Q.fasta -task blastn -max_hsps 1 | grep LC074724
$
WTF?
I’ve been using BLAST for the last 4 years or so, and have only today realized that something like this could occur.
It’s of course super important.
And despite all the above, I still don’t understand what’s going on.
Here’s a gist that discusses what looks like the same issue. I pasted the above into it.
A blog post on bugs in BLAST and the difficulty of filing them, getting them fixed, etc.
See also this post in which the user adds the -max_target_seqs option (setting it to 10) and the the top hit vanishes. That’s the opposite to my case, where adding the option (with a high value) causes a previously very-high-bitscore-but-unmatched (or unshown) sequence to be matched (or shown).
Do stuff on things, in parallel
18:10 August 5th, 2017 by terry. Posted under me. | 2 Comments »
Most people don’t really know how to use the tools they spend the most time using: the shell and their editor. It’s worth stopping once in a while to teach yourself new things. Sometimes you find something so powerful that it has a huge impact, in which case it’s even worth stopping other people so they can maybe learn it too.
GNU parallel is such a tool. Actually, I think it’s the most powerful shell utility I’ve ever used. I heard about it when it first came out (in 2011), read the manual, thought “amazing!” but immediately went back to using xargs (I thought I was too busy).
Often I find I need to be exposed to these tools multiple times before I really pick them up and use them regularly. Try to find one thing that it’s useful for you and to start using it just for that. But keep in mind what else it can do and gradually expand your usage. Read the manual repeatedly (over the years). Write down a few options or command lines and stick the piece of paper near your monitor. Or just go to talks by geeky friends.
Things
You very often need to take some action on a collection of things. E.g., list a set of files.
How do we get the list of things to operate on? The shell has always provided some help, and there are various standalone tools that can help.
Globbing
The shell gave you globbing
# List all files ending in .c
$ ls *.c
# List all files ending in .c or .h
$ ls *.[ch]
and glob made its way into programming languages
# Python
from glob import glob
print(glob('*.c'))
Making things up: echo
echo is extremely useful. It’s like print in a typical programming language.
$ echo a b c
Making things up: brace expansion
In bash (and some other shells), brace expansion is very useful
$ echo {chair,stool,table}.c
chair.c stool.c table.c
Note the important difference from globbing: the things don’t have to already exist as files or directories. Brace expansion is just creating strings while globbing is expanding patterns into the names of pre-existing things (file and directory names).
When used more than once, brace expansion gets you the cross product:
$ echo {chair,stool,table}.{c,h}
chair.c stool.c table.c chair.h stool.h table.h
This is (roughly) just a loop in a loop.
Command expansion
If you put a command in $(...), the shell runs the command and replaces the whole expression with the output of the command. So this
$ wc -l $(grep -l hello *.c)
runs wc -l on the .c files that contain hello.
Finding things
It often becomes awkward to use globbing, so there’s a separate find command that can find things (files or directories).
# Recursively find files whose names end in .c and print their names.
$ find . -type f -name '*.c' -print
You can’t do that with globbing, unless you use
$ ls *.c */*.c */*/*.c */*/*/*.c
which has obvious limitations.
Apart from being able to walk the filesystem, find has many options to only return files (or directories) with certain properties.
For example, here we find files whose names contain ‘abc’, with a size of over 1MB and that have been modified in the last 2 weeks:
$ find . -type f -name '*abc*' -mtime -2w -size +1MB
Warning: find is also a bit cryptic. I still don’t really understand it, after over 30 years!
Altering things
It’s very common that you need to change the names of things slightly. There are many standard UNIX tools that can help: tr, cut, basename, dirname, sed, awk, perl. It’s worth learning very basic usage of these things (especially tr and cut). See next sections for some simple examples.
Do stuff on things
Shell variables and loops
Many people do not realize that the shell is a programming language. It has variables and loops, which you can use to build up a list of things:
$ for year in 2015 2016 2017
do
for name in sally jack sue
do
mkdir -p $year/$name
done
done
The above could be done with brace expansion:
$ mkdir -p {2015,2016,2017}/{sally,jack,sue}
Often you use the value of a variable to make a file with a related name. Use command expansion $(...) plus one of the above altering tools to make the new name:
$ for file in *.c
do
base=$(echo $file | cut -f1 -d.)
wc -l < $file > $base.line-count
done
Using exec in find
Find offered its own limited way to run commands on things it found. E.g.,
# Find .c files and run wc on each of them.
$ find . -name '*.c' -type f -exec 'wc {};'
This was quite limited and it results in wc being run once for each file, which is much slower.
Enter xargs
If you’ve never learned xargs, don’t bother with it, just skip to parallel (see next section).
To complement find, xargs came along, reading a list of things from standard input:
$ find . -name '*.c' | xargs wc
By default, xargs takes all the names on standard input (splitting on whitespace and newline) and puts them at the end of the command you give it.
A big difference in the above is that wc is (normally) only run once.
xargs can also make sure that the command line isn’t too long (it will invoke wc more than once if so) and can be told to only give a certain number of things to a command.
This will give 5 things at a time to wc:
$ find . -name '*.c' | xargs -n 5 wc
You can also tell xargs where to put the things in the command
$ mkdir /tmp/c-files
$ find . -name '*.c' | xargs -I+ mv + /tmp/c-files
Or
$ echo a b c d e | xargs -n 2 -I+ echo mv + /tmp
mv a b /tmp
mv c d /tmp
mv e /tmp
xargs will run into problems if argument names (usually file names) have spaces or newlines in them. So find and xargs can use the same convention to NUL-separate names:
$ find . -name '*.c' -print0 | xargs -0 wc
This is the accepted / standard safe way to use find & xargs.
Do stuff on things, in parallel
GNU parallel
This was all a bit shit. It was hacky, there were exceptions, there were limitations, there were conflicting versions of programs (e.g., OS X xargs is crappy compared to the Linux version). You could do lots of stuff, and it felt powerful, but you’d often end up writing a shell script if you had to do something slightly different (like make a new file whose name was based on a simple transformation of another file’s name):
$ for file in *.c
do
base=$(echo $file | cut -f1 -d.)
wc -l < $file > $base.line-count
done
And, when you had your loops and your find and xargs all just so, your commands were still executed one by one. So there you are, on a machine with 8 cores but you’re only using one of them. It’s no big deal if your command is trivial, but if takes an hour, you might be looking at an 8 hour wait instead of a 1 hour one.
This is not so easily solved. You could do something like this:
$ for file in *.c
do
wc $file &
done
but that runs all your commands at once, with no regard for how many cores you actually have. That can be even worse than just running one command after another. What you in fact want is one command running on each core, with a queue of pending commands that are started as cores become free.
GNU parallel solves all these problems. It gives you looping, can read input in words or complete lines, has powerful ways to use and manipulate the names it is given, does things in parallel (but can still order its output to match the input). It can even send jobs to remote machines.
Let’s have a look.
Emulating xargs
You can use parallel in place of xargs. Here’s the setup:
$ mkdir /tmp/test
$ cd /tmp/test
$ touch a b c
$ ls -l
total 0
-rw-r--r-- 1 terry wheel 0 Aug 5 17:09 a
-rw-r--r-- 1 terry wheel 0 Aug 5 17:09 b
-rw-r--r-- 1 terry wheel 0 Aug 5 17:09 c
The following passes each file name to echo individually:
$ ls | parallel echo
a
b
c
Whereas this collects the multiple names and puts them all to one invocation of echo:
$ ls | parallel --xargs echo
a b c
Note that this is subtly different from the following:
$ echo * | parallel echo
a b c
$ echo * | parallel --xargs echo
a b c
That’s because ls will write one filename per line of output when it detects that its stdout is not a terminal (contrast what you get when you run $ ls with ls | cat). On the other hand, echo * writes just one line of output.
In both the latter (echo) examples, parallel is just getting one line of input and is giving that line to echo. In the former case (ls) it gets multiple lines of input and the --xargs option tells it to collect those lines and put them on the command line to echo.
Note that I don’t understand why parallel -m and parallel -X don’t also collect input lines in the way --xargs does. The manual page for parallel seems to indicate that they should.
Sending commands into parallel
$ for year in 2015 2016 2017
do
for name in sally jack sue
do
echo mkdir -p $year/$name
done
done | parallel
parallel can make loops for you
Here’s a cross product loop, just like the above:
$ parallel echo mkdir -p '{1}/{2}' ::: 2015 2016 2017 ::: sally jack sue
Output ordering
Because processes may not finish in the order they’re started:
$ parallel echo ::: $(seq 1 10)
7
8
9
6
5
4
10
3
2
1
there’s a -k option to make sure the output order matches the input:
$ parallel -k echo ::: $(seq 1 20)
1
2
3
4
5
6
7
8
9
10
Reading the names of things from files
$ cat names
sally
jack
sue
$ parallel echo mkdir -p '{1}/{2}' ::: 2015 2016 2017 :::: names
And
$ cat years
2015
2016
2017
$ parallel echo mkdir -p '{1}/{2}' :::: years :::: names
Or read from standard input, from a file, and from the command line:
$ ls *.c | parallel echo '{1} {2} {3}' ::: - ::: years :::: names
Combining input names
Sometimes you don’t want a cross product, you want to combine names (like using zip(...) in Python or (mapcar #'list ...) in lisp to combine multiple lists). Compare
$ parallel echo '{1} {2}' ::: 2015 2016 2017 ::: goat monkey rooster
2015 goat
2016 goat
2016 monkey
2015 rooster
2015 monkey
2016 rooster
2017 goat
2017 monkey
2017 rooster
with
$ parallel echo '{1} {2}' ::: 2015 2016 2017 :::+ goat monkey rooster
2015 goat
2016 monkey
2017 rooster
Modifying names
Parallel has a bunch of ways to edit names. So instead of needing to write a script like this:
$ for file in *.c
do
base=$(echo $file | cut -f1 -d.)
wc -l < $file > $base.line-count
done
you can just do this:
$ parallel 'wc -l {} > {.}' ::: *.c
There are lots of ways to modify input names, including:
- {} The name (i.e., the input line), unmodified
- {.} Input line without extension.
- {/} Basename of input line. E.g.,
/home/pete/main.cbecomesmain.c. - {//} Dirname of input line. E.g.,
/home/pete/main.cbecomes/home/pete. - {/.} Basename of input line without extension. E.g.,
/home/pete/main.cbecomesmain.
And the --plus option gives you more, like {..} to remove two dotted suffixes.
You can modify input names individually:
$ parallel echo '{1/} {2.}' ::: data/2015 data/2016 data/2017 ::: sally.c jack.c sue.c
2015 sally
2015 jack
2015 sue
2016 sally
2016 jack
2016 sue
2017 sally
2017 jack
2017 sue
Running on remote machines
The following transfers all *.c files to a remote machine ac, runs wc -l on them (one by one), puts the output into a file that has the .c replaced by .out, returns all the output files to my local machine, and cleans up the files created on the remote:
$ parallel -S ac --transferfile '{}' --return '{.}.out' --cleanup wc -l '{}' \> '{.}'.out ::: *.c
The option combination --transferfile '{}' --return --cleanup '{.}.out' is so common you can abbreviate it to --trc '{}.out'.
$ parallel -S ac --trc '{}.out' wc -l '{}' \> '{.}'.out ::: *.c
A real life example
Here’s a script I wrote
sample=`/bin/pwd | tr / '\012' | egrep 'DA[0-9]+'`
# Collect all read ids, with > replaced by @
cat 03-panel/out/[0-9]*.fasta | egrep '^>' | sed -e 's/^>/@/' > read-ids
count=1
for dir in ../../2016*/Sample_ESW_*${sample}_*
do
fastq=
for file in $dir/03-find-unmapped/*-unmapped.fastq.gz
do
fastq="$fastq $file"
done
# Pull the FASTQ out for the read ids.
zcat $fastq | fgrep -f read-ids -A 3 | egrep -v -e '^--$' | gzip > run-$count.fastq.gz
count=`expr $count + 1`
done
I made it faster by running a zcat on each core using parallel:
# Some lines omitted
for dir in ../../2016*/Sample_ESW_*${sample}_*
do
fastq=
for file in $dir/03-find-unmapped/*-unmapped.fastq.gz
do
fastq="$fastq $file"
done
ls $fastq | parallel "(zcat {} | fgrep -f read-ids -A 3 | egrep -v -e '^--$')" | gzip > run-$count.fastq.gz
done
But that’s still inefficient because my main loop waits until each dir is completely processed (i.e., all its fastq files have been run). So as the last fastq files are being processed, cores are unused.
So, faster:
# Some lines omitted
for dir in ../../2016*/Sample_ESW_*${sample}_*
do
fastq=
for file in $dir/03-find-unmapped/*-unmapped.fastq.gz
do
fastq="$fastq $file"
done
echo "zcat $fastq | fgrep -f read-ids -A 3 | egrep -v -e '^--\$' | gzip > run-$count.fastq.gz"
done | parallel
This uses the fact that parallel will treat its input lines as commands to run (in parallel) if it’s not given an explicit command to run.
And this could have been made faster by using parallel to run the zcat.
Counting sequences in FASTQ
I wanted to count the number of nucelotide sequences (billions of them) spread over nearly 6000 FASTQ files (found under directories that start with 20):
$ find 20* -maxdepth 2 -name '*.fastq.gz' | parallel --plus --bar "zcat {} | egrep -c '^\\+\$' > {..}.read-count"
The --bar gives a cool progress bar. The --plus makes {..} work (to remove two suffixes).
And there’s much more
- Breaking input up by delimiter instead of by line.
- Breaking up input into chunks and passing each chunk to a process that reads from
stdin(using--pipeor--pipepart). - Stop launching jobs after one (or a percent) fail. E.g.,
parallel --halt now,fail=1 - Kill currently running jobs if one fails.
- Resource limiting.
- Resuming failed jobs.
- Retrying failing commands.
- Dry run:
--dry-run. - Using
tmuxto show output.
Installing
On OS X, if you’re using brew:
$ brew install parallel
More info
- Run
man parallel(see many examples at bottom). - Parallel tutorial.
- Wikipedia page.
- Some videos.
Thoughts ahead of the 2017 Transcontinental Race
04:14 July 25th, 2017 by terry. Posted under me, travel. | Comments Off on Thoughts ahead of the 2017 Transcontinental Race
 Last Sunday I did my final training ride before the Transcontinental Race (TCR), which starts in Geraadsbergen (Belgium) at 10pm this Friday night, July 28, 2017.
Last Sunday I did my final training ride before the Transcontinental Race (TCR), which starts in Geraadsbergen (Belgium) at 10pm this Friday night, July 28, 2017.
In February 2016 I happened across Joe Todd’s extraordinary TCR blog. I was captivated, and stunned to learn that people went on 4,000km cycling races across Europe, riding with very little sleep for two weeks, carrying what looked like almost nothing (see the first photo on Joe’s page). It seemed incredible. I instantly knew that I had to do it too, or at least try.
I couldn’t really sleep for the next two nights. I couldn’t stop thinking about the TCR. Just the name, the Transcontinental, just the sound and the idea of it, was irresistible. It somehow conjured exotic youthful thoughts of the impossibly distant Trans-Siberian Highway, Murder on the Orient Express, etc. The more I read about the race, the better it seemed. I loved the spirit of the event, the honor code, the fact that the riders have a say in what is unsupported (1, 2), the inclusiveness, the lack of rules, the madness of it, and the rawness. The distances these people were covering every day seemed crazy. And they were doing it in the mountains, with bags on their bikes, in snowstorms and baking heat. Getting blown off their bikes. Getting lost. Barely sleeping. Totally unsupported. Totally nuts.
And glorious:
How could you not want to be a part of that? To have that be a part of you?
Entries for 2016 had closed, but I mailed the organizers, Mike Hall and Anna Haslock, to ask if I could possibly still get in. They replied to say no, but that if I volunteered to help on the 2016 edition that I’d have a guaranteed place in 2017. I ended up building them a web site with a map to show information about hundreds of Eastern European border crossings, allowing people to submit comments on them. The site wasn’t really used in the 2016 race as there was just too much going on for the organizers ahead of the race. I got to hang out on Slack with the admin team and got some glimpses into what goes on behind the scenes both ahead of time and during the race. One thing I realized is that if you race the TCR you might (very often) feel like you’re all alone, but in fact you’re not. There are dozens of volunteer “dot watchers” spread around the world, who monitor the rider tracking map around the clock. The dot watchers are dedicated and experienced, and they really care. Every rider is closely monitored. The team of dot watchers are still there, watching, weeks after the first rider finishes the race.
I doubt that a single day has passed in the last 18 months without me thinking about the TCR. And I don’t mean having the odd passing thought once or twice a day. I mean thinking about it all the time. For the last six months, at least, when I wake up in the morning the TCR is in my head within the first minute. The TCR rushes in to fill any idle time in my day. I go to bed thinking about it. It has become an obsession. There’s no better word. I read everything I could find online, slowly accumulating knowledge of people’s experiences, equipment, choices, and so on.
I began to ride longer distances, with more challenges. There were questions and uncertainties that could only be resolved on the road. In 1989, aged 25, I’d cycled from Munich to Madrid, 2100km in a month, happily sleeping by the road, in sheds, and on building sites. That worked out at about 100km a day, with a week of rest days. A couple of years ago I thought I was unlikely to ever regularly do rides of that length again. But I started going out on Sunday club rides with the local St Ives Cycling Club, and so began to regularly ride over 100km.
Last year I did an Everesting. I rode to Rotterdam and back a couple of times. I rode to Bonn and back. In 2017 I’ve been going after more distance and more climbing. I’ve been hill-climbing in Wales in the ValleyCat, to Yorkshire, in the Cotswolds, in Scotland, and in the Peak District. Most recently, with David, I did a 430km loop including the Dunwich Dynamo. The daily distances have slowly gone up… 150km, 200km, 300km, 400km. The amount of climbing per day has gone up… 2000m, 3000m, 6000m, and of course the Everesting was 8900m (but was relatively very easy climbing). I’ve learned to ride more slowly, conserving energy, riding for 10, 12, …, 16 hours, combined with some resting, and lots of eating and drinking. I raced in the Tour of Cambridgeshire in 2015, in what seemed like a fast time but which now looks slow. I religiously followed the British Cycling Advanced training plan for 3 months at the start of 2016, riding in the rain and the snow, come what may. Then I did the Tour of Cambridgeshire at a much higher pace in 2016, and 2017. I qualified for the UCI Gran Fondo world championships each year, and even went to Denmark to race it (slowly!) in 2015, representing Australia. In 2016 and 2017 (so far), I’ve covered 22,000km. For the last 7 months I’ve been working out 2 or 3 times a week, doing weights, core exercises, and stretches. I taught myself something about bicycle maintenance. I got a fantastic new MASON Bokeh bike, at great expense.
Lately I’ve spent tons of time planning the 4000km route. It sometimes feels like a full-time night job. I’ve been over at least double that distance, looking at route alternatives. There are so many tools to help with the job, and yet many (small) sections of the route are unknowable, at least from this great remove. Things will undoubtedly go wrong. They always do. But I’ve spent a ton of time trying to keep them to a minimum. In case you’re curious, here’s what I’ll be packing (still to be pared down a little).
Anyway now, finally, the 2017 TCR is right around the corner!
Here’s the 2017 teaser video, which I wish was a little longer:
This Thursday (July 27, 2017) Derek or David will drive me to Belgium for the start at Geraadsbergen. Here’s a video of the TCR start in 2016. At 10pm 300 riders, more or less, will head off into the night, and I’m so happy I’ll be one of them. Here’s an article giving a bit of an overview of the route.
There are four checkpoints you have to ride through, each with a mandatory parcours section. One is the famous Semonzo ascent of Monte Grappa. Another is the (infamous) Transfăgărășan highway in Romania:
I just found out that the Transfăgărășan is closed to cars from 9pm to 7am each night, though I’ve no idea why. Someone posted that info to Facebook, to which there was this reply:
Bagoly Levente: This is only for motorised traffic. I know cyclists who passed it this weekend from midnight till four in the morning till vidra dam with no problem.
But its quite scary in the dark. Keep in mind that once your up it doesnt mean that your doing 50km/h till the dam. Besides the lake there is many ups and downs which youll climb slowly and after darknes sets it very scary and dangerous because of people leaving garbage all around the place and bears do come down to check it out. I was passing through there around 10pm after a 600km endurance ride and i have to say i almost layed some bricks along the way… Its a good thing if you have a wistle like i did and blow it every now and then. pepper spray wont help against wolves or bears. although between these two only the wolves will hunt on you. bears only attack under few circumstances when you surprise them. thats why a wistlesound which travels dar away will give them a signal and they will get out of your way. bears dont want any trouble. wolves do.
Hopefully I’ll pass through there in the daylight…
If you’d like to follow along, you’ll be able to track riders at either TrackLeaders or FreeRoute. Here’s the rider list; I’m #85. There’s a party in Meteora (Greece) on August 12, which is day 15. To make it to the party you need to average about 266km per day. I think I can do that, but it’s impossible to know. The distance itself would be hard if that was all there was, but my route has about 37km of climbing, and parts of the route will be extremely hot (over 40C / 104F).
Because the race is strictly unsupported, you can’t have any form of help that wouldn’t be available to all riders. So you obviously can’t have friends sending you SMS messages telling you how to get un-lost or making you hotel bookings, or… anything really.
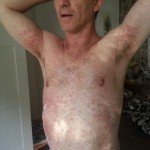
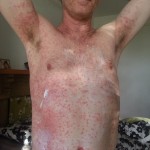
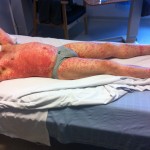
But you can help by sending encouragement during the race. I’m sure I’ll have lots of fun, but it’s also going to be utterly exhausting. By far the physically hardest thing I’ll have ever attempted. Just look at Alexandre Bourgeonnier’s face in this picture taken as he finished the 2015 race.
So please feel free to send some friendly words in the middle of the night, or any time. I gave up on all social media some years ago, but have recently been hanging out on Facebook because there’s a Transcontinental group there, with tons of other TCR racers and ex-racers, and information. So I guess I’ll be posting things on FB. I’ll post progress sections to Strava. I also have a Twitter account that I haven’t used in nearly 5 years and an Instagram one too (similar) – neither of which is likely to get used, but who knows? You can always send me email or SMS. I probably wont be quick to reply, though. I will likely have my phone in aeroplane mode for much of the time (to save battery), so don’t panic if you try to call and you get voicemail or I don’t answer. Also, people’s dots frequently stop moving on the map for a wide variety of reasons, so don’t panic over that either. I stop often. Too often. It’s a bad habit I’m hoping to greatly improve on.
Want to read more about the TCR? You could read more of Joe Todd’s blog. There are many other write ups of people’s experiences here. I’ve read them all. Many are highly amusing. Or just search online for TCR accounts and videos – there’s a ton of them. Chris White, a TCR veteran, has put together a detailed, comprehensive, and authoritative overview of long-distance cycling in general and the TCR specifically, at ridefar.info.
Here’s an article from a few days ago with an opening paragraph I can relate to:
For the last I don’t know how long there’s been a dull undercurrent of fear, panic and restlessness. It’s that pre-big-exam dread combined with the giddy stomach waltzer of falling in love. A feeling that’s a sticky apprehension churned together with an itching excitement to create a thick cloying inability to be able to think about anything else. A little bit sick all the time, careering in jolts between abject panic and serene calm without touching anything in-between. And then back again. In minutes.
I don’t know if I’ll complete the TCR. I don’t think anyone can know, because there are so many things that can go wrong. People get achilles and knee problems. They get nerve damage in their hands and can’t even hold a knife and fork to eat. They get Shermer’s Neck, back problems, and occasionally food poisoning. Bicycles break. And of course, there are accidents, sometimes tragic.
Here’s a video about the 2016 race:
#171 The Transcontinental Race journey from PEdALED on Vimeo.
Over the last year I’ve tried not to let the talking get ahead of the reality. But now I’m finally there, and about all that’s left to do is to start the thing. I’ve gone from naïvely thinking that of course I could do it (2016), to more realistic thinking there’s no way I could (early 2017), to now being cautiously optimistic that I’ll be fine. I could be fitter and stronger, and I have a couple of niggles (hip, hamstring) that I hope will dissipate, but overall I’m happy with where I am and my level of fitness and confidence. You simply cannot know how it will go, though. Mentally, I don’t know what will happen. I’ve never needed much sleep, but that’s under normal conditions. Maybe I’ll race madly and barely sleep. Maybe I’ll be so exhausted I’ll just give up on the racing and aim to finish by the party. Maybe I’ll give up altogether (though I bloody hope not!). All I can confidently say is that the things I have control over appear to be under control. Now I just have to avoid the wild dog pack attacks in Eastern Europe and Greece, not to mention the wolves and bears in Romania.
Here’s my bike, fully loaded. It weighs 19kg (41.8lbs) with no water or food! Cycling up steep hills or mountains is hard enough even without any bags :-)

And the elevation profile of my route:
![]()
THANKS so much to Derek Smith, David Pattinson, Chris Lloyd (of Chris’ Bikes), Duncan Chapman (calves), Arwen Altenberg (wardrobe), Chris White (wheel), Sarah Kelman (weather), Josh, Sam, and Andy at Bicycle Ambulance, Dom Mason and Cal Nicklin at MASON (bike, clothes), Steve Lindley (lock), the friendly members of the FB TCR group, Supernova Lights (40% TCR rider discount), Bethan Roderick and Dafyd (staying up til 3am to feed me after the ValleyCat), Tom Kirkpatrick and the TCR organizational team, the SICC riders who taught me how to ride a bike, and finally to Anna Haslock for her bravery, and the amazing Mike Hall who created the TCR, and who I unfortunately never got to meet.
Everesting
03:42 September 5th, 2016 by terry. Posted under me. | 11 Comments »
Yesterday (Sept 3, 2016) I did an Everesting ride on my bike. As a concept, Everesting is dead simple. You pick a hill, any hill, and ride up it enough times to equal or exceed the vertical height gain of mount Everest (8,848 meters). Here are the rules. There’s a Strava blog post, The Ultimate Climbing Challenge: Everesting, that gives a good overview and some history.
I’d read a bunch of other people’s blogs about Everesting and found them helpful, so I’m hoping my own write up will help others. Also I know that if I write it all down I’ll come back and read it over the years, be happy I took the time to do it, and that I’ll forget almost all the details if I don’t. I decided to be selfish and write for myself, so if you’re looking for Everesting advice you’ll have to bear with the prose. But that’s ok, because if you can’t handle reading a long web page you might not be able to handle an Everesting anyway :-)
There are lots of Strava links below that you wont be able to follow unless you have (or create) an account, sorry.
Cast of characters
- Derek Smith – my partner in crime, who organized things, brought a ton of supplies, drove us out there, stayed the whole time, and rode about 20 ascents on a bike with highly unsuitable gearing.
- James Barlow – a St Ives CC cyclist who started the attempt but had shoulder and neck problems half way through. James is a much better, much stronger, and much faster cyclist than I am.
- David Pattinson – my office mate, who swooped in unannounced to ride the last 20% with me.
- Beth Evans – just graduated Cambridge and has been working with us over the summer. A British Military Fitness enthusiast. Don’t mess with Beth.
- Jeremy OneLung – St Neots based cyclist who probably holds the record for most number of guided busway rides.
- Ana, Lucas, and Findus – my wife and our two youngest kids.
Before anything else, THANKS to everyone. Though I perhaps could have done it without you, I really don’t think I would have. I’d have been too wet and cold and miserable to go on. It ended as a team effort, as you’ll see.
10% of an Everesting
I think David first told me about Everesting, and we said how silly and extreme it sounded, and of course said many times that we’d like to do it.
I’ve been doing quite a lot of cycling training this year, including trying to get stronger on hills. Unfortunately there really are no hills around here (in Cambridgeshire). But there are some climbs that are still challenging if you’re not much of a climber (like me) and you try to do them in a difficult way (e.g., in a very high gear, or standing up on the pedals, etc). I’d been happy to feel myself getting stronger on hills and to reach the point where I was able to stand on the pedals climbing for about 4 minutes.
I was in Holland a few weeks ago, cycling with two of my kids. We had a rest day in Nijmegen, where there are some small and a bit steep hills rising from the river into the city. I decided to go back to a ramp I’d ridden up with the kids and do some tiny intervals on it. Later, Derek took some screen shots from Google street view, one from the bottom, one at the top looking back towards the city:
I took my son’s bike, which is my old Boardman Hybrid Pro. Real road cyclists would turn up their noses at it, but it’s a damned good bike, not much different from a road bike (just the handlebars and a little heavier). I did a few ascents, standing up and in a pretty high gear to make it hard on my legs. After 10 ascents or so, I started to think about how many I should do. The ramp is only about 100m long and according to my Garmin had about a 9m altitude gain. So it’s something like a 9% average incline. I decided I could probably manage 100 ascents, and that’s what I did. I did the laps all standing up, though I did click down to a slightly easier gear. People looked at me as though I was a bit odd, and asked questions in Dutch.
At the end I wasn’t tired. Or maybe I was, because I titled the ride “1% of an Everesting”. I thought it wasn’t so bad, and that maybe, just maybe, I could do that 100 times. Derek then pointed out that it was actually 10% of an Everesting! At that point I thought wow, on the right hill and with my road bike, I could definitely do an Everesting. The 10% took 1.5 hours, so maybe 15 hours for the full thing.
My Garmin (or GPS) went nuts on the ride – a recurring theme, as you’ll see – and showed that I’d made a quick side trip to somewhere hundreds of kilometers away, at an average speed of about 200km/h. So I reluctantly deleted the ride and added a manual entry instead. James made a comment, asking if I wanted to do “something silly” – a full Everesting, and I said yes. Jeremy suggested I edit the original Garmin file to crop out the spurious extra hundreds of kilometers. I used Fit File Tools to do that and it worked out perfectly.
At that point, an Everesting attempt with James was on. We just had to find a hill and pick a date.
Finding a hill
I wanted to do the Everesting in Cambridgeshire, because that’s where we live and there hadn’t been an Everesting done here – perhaps because there are almost no hills, and certainly no big ones.
James suggested either the weekend of 27/28 of August or the weekend of 3/4 September. I mailed Derek and David, who both replied that they’d be out of town the first of those options but back on the 3rd/4th. Derek said he’d do all the logistics and ride a bit, and David said he might join and would at least come along for part of it.
James suggested we use The Heydon Mountain Top Ascent of Gran Canaria. It’s a 2.2km segment and you gain 80m. It sounded good at first. But the Everesting Cycling Calculator said we’d have to do 107 laps to gain the required altitude, a ride of 480km! James has cycled nearly 17,000km already this year, and can do that, as he’s proven repeatedly. But could I handle nearly 500km in a day, with most of the time spent going uphill? I doubted it. Derek and I did a recon ride to check it out. Apart from finding the segment too long, the road was a bit rough and it felt to me like there would be too much thinking involved. I’d read multiple blog posts about people going to “a dark place” and hallucinating after 7,000m of climbing. I didn’t want to be doing any thinking by that point, and the segment had some curves you can’t see around, a high-speed curve in the descent, an awkward corner at the bottom turnaround point, and seemed a bit too populated at the top.
I told James I’d do it but that I wanted to keep looking to see if there was a better and shorter option. That therefore meant something steeper. But where? Velo Richard has written a couple of blog posts, The ahem Big Cambridgeshire Climbs and The Short and Steep Cambridgeshire Climbs which should give you some idea of the “hills” in this part of the world. I picked out two possibilities, very close to each other.
The first was the Barham climb, in which you gain 13m over 200m, requiring 707(!) laps, but needing a total ride distance of only 300km. I went out and rode it and didn’t like it one bit. It’s a tough little climb, probably over 10%, on a rough surface, with no visibility at the top for oncoming traffic.
The second possibility was Staunch Hill. I went and rode it half a dozen times and found it almost perfect. It would be 247 laps, for a total of 280km riding. That was definitely in my distance range, though I didn’t know if I could handle that much hill climbing.
20% of an Everesting on Staunch Hill
So I went out a few days later and did 20% of an Everesting on Staunch Hill. I did it party in daylight, partly in the dark. It was great, really great, and I knew we had our hill. Or at least I knew I had my hill, and was hoping James would be happy with it too. And he was. It took me almost exactly 3 hours to do the 20%, which was right on line with the 10% in 1.5 hours in Nijmegen.
What made Staunch Hill perfect? It’s dead straight. Although you can get to it from the A14 highway, you can only do that from one direction and the road only leads to a tiny quiet village, Leighton Bromswold (population about 200). The surface was smooth and there were no potholes at all, just a couple of very minor bumps that you could easily ride over with no problem. The road was wide enough for 2 cars to pass. At the top there was a spacious and open T intersection with essentially zero traffic. And a nice-looking pub, The Green Man (first licensed in 1650), which I told myself it would be great to eat in and have a final beer in, even though I doubted I’d actually do that. At the bottom there was tons of visibility in both directions. The bottom was a little narrow, but not so narrow you needed to unclip. In the 3 hours I was riding I was only passed by about 10 cars, all driving carefully and not at high speeds.
It also seemed perfect in terms of what I needed to do to get up it. My bike has a compact (50/34 teeth) chainring on the front, and Derek and I had (eventually) put an 11×32 cassette on the back when I was getting ready to do a ride in the Peak District. I have a Stages power meter which, as a not-very-experienced cyclist, I find incredibly useful. I’ve been using it for maybe 6 months now, and I have a pretty good idea of what power levels I can attain and (more importantly) sustain. I rode Staunch Hill very carefully, watching the power meter the whole way. I found that if I was on the small ring (34 teeth) and the 32 tooth rear cog, and kept the ascent as gentle and easy as possible, that I could keep peak power down to 280 watts. My Functional Threshold Power is about 250W (i.e., I can in theory sustain 250W for an hour). I figured I could handle pushing up to 280W briefly over and over again, as long as I got a rest in between. BTW, 250W is not very much – it’s probably what you use in one leg just climbing stairs. If you’ve ever gotten up on your feet out of your saddle to go up any incline on a bike, you’ll likely have exceeded 280W.
I also know a lot about my heart rate, again from doing lots of training. On Staunch Hill I was peaking at about 155bpm, well below what I can sustain for an hour (about 170bpm).
So I was fairly confident that if I just made sure to ride slowly and at those power and heart numbers that I could probably go on for a long time.
Back to the story… we now had a date and a hill and 2 weeks to wait.
I did a 113km ride the day after the 20% ride, and then the 203km Mildenhall audax the next weekend. In the week before the Everesting I took it very easy, just cycling into Cambridge and back once (under 40km in total).
The spreadsheet
It would be wrong to leave Derek’s spreadsheet out of the story.
He made a spreadsheet on Google docs and shared it with David and me. I took a look and wanted to run! It seemed so over the top, with 60 rows of stuff. Included were 10 spare tubes, 2 spare tires, an extra bicycle, a bike maintenance stand, and many other things, some with eye-popping quantities. I couldn’t handle it, and decided to politely ignore the spreadsheet entirely. David took a different approach and added items Derek had forgotten: crimpons, rope, compass, carabiners, ice pick, oxygen tank, and sherpa.
As we got closer to the day, I thought better of my silence and discretely applied strikeout to about half the items on the sheet and replaced some of the quantities by their log base 10. We still had enough equipment and supplies on the day to support a small team of Everesters.
Here’s a screenshot of just the lower part of the Everesting spreadsheet (click to see the details):
To be completely fair, I added (and then crossed out) the broom, which I thought we might use to clear loose gravel or stones from the sides of the road in the narrow turnaround area.
Linear scaling?
Although I was feeling pretty confident, there was an unknown lurking right at the heart of all pre-Everesting discussions, thought, and planning. How would things scale for me over time? I.e., was it safe to assume that the last 80% of the ride would be just like the 20% that I had actually done? Or would things go non-linear? I’d read many blog posts talking about hitting the wall at about 7,000m, etc. Derek and I could only guess at what might happen to me, we really didn’t know. So of course we had to be on the safe side. I’d done the 200km ride in 6 hours 45 mins, on just a piece of cake, a sandwich, a candy bar, a coffee, and several bottles of water. It was obviously not safe to just double those numbers and hope for the best. And what about my heart rate over the ride? Would I have cardiac creep, in which your heart rate goes up even though the power you needed to input stays flat? If so, I would be in trouble because it hadn’t been possible to get up the hill in my lowest gear at less than a 280W peak. What about my body – my knees, my neck and back, my muscles, other joints? I just didn’t know what would happen, and wished this wasn’t my first Everesting.
100% of an Everesting
Derek volunteered to drive me out there, with both our bikes in the back of his car. He said he’d stay through the whole thing. I thought that was unnecessary and asked him what he planned to do that whole time, especially sitting there alone in his car at the top of the hill for many hours in the dark. He told me he was planning to monitor my nutrition – carbohydrate, protein, liquid consumption, etc. I wondered how he was planning to do that, and told him that I felt like some kind of laboratory animal in an experiment. Anyway, I was very glad for the company. If blogs about Everesting agree on any one thing, it’s the need for good support, and I definitely had it.
We’d arranged to meet James at the top of Staunch Hill at 1am on Sept 3, 2016. He’d suggested the night before that maybe we could meet at 8pm to avoid the predicted rain in the afternoon of the 3rd, but I wouldn’t have been able to sleep before the ride. It didn’t make sense to me to do an Everesting without sleeping beforehand! But James is good at finding ways to make rides more extreme, like adding 225km round trip to get to & from a 175km sportive, and probably didn’t think that was a big deal. When I told him I’d like to sleep, he said yes, that I could do plenty of sleeping after the Everesting. I told him to go for it if he wanted, but that I much preferred the sleep plus rain combination (which is exactly what I got). In the end we settled on a 1am start. Derek and I drove out there, while James did a little 27km warm up ride from St Ives, including 300m of pre-Everesting climbing.
We arrived about 15 minutes ahead of James and I changed into my gear and got ready. I asked Derek if he was going to ride some laps with me, and he said he thought he’d better not because he’d have to use a light and it would be better to keep his light in reserve for me. That turned out to be code for “I’m about to have a 7 hour sleep in the back of my car.”
I did a few laps and then James arrived. He said he was almost ready to begin, but needed a cigarette first. I’m not making this up. Derek suggested that the Hell’s 500 people who maintain the Hall of Fame could add an asterisk next to his name as someone who smoked during the Everesting. I hate to think what his cycling would be like if he didn’t smoke! It subsequently occurred to me that perhaps James doesn’t actually smoke! That maybe he turns up at ultra distance events, and while everyone else is stretching, slamming last-minute gels and organic carb and ethically-sourced protein bars and nervously eyeing other cyclists, James says “Hang on a minute guys, I need a final smoke.” At that point the event may as well be over. Stephen Potter, who wrote a classic little text on gamesmanship that I read when I was about 14, would have been deeply impressed.
Turn off your mind, relax and float downstream
I got into a rhythm, doing each lap in almost exactly the same way. Descend completely relaxed, with no pedaling but stretching my legs a bit. Brake and turn at the bottom after looking over my right shoulder for cars. Keep the first (quite flat) part of the ascent at under 200W. Begin changing to lower gears at the sign on the right. Be in the 28 tooth cog just after the dark patch on the road. Be in the 32 tooth just after the strip of lighter-colored concrete a bit later. Ride up as gently as possible, eyes on the power meter. Keep it under 280W when it got steepest. Breathe properly. Relax. Hands loose on the handlebars. Stand up on the pedals on the flat at the top and stretch both legs. Push the lap button on the Garmin at the same place at the top every single time. Stand up and do a few seconds of mini sprint on the flat heading back towards the descent. Change down 4 gears so I wouldn’t have to change at the bottom. Repeat. That was basically it for 16 hours of riding.
A few songs went round and round in my head. In particular, The 801‘s beautiful 1976 cover of Tomorrow Never Knows (which I somehow didn’t even realize was a cover, even though I had heard the Beatles version many times). Lyrics.
Turn off your mind, relax and float downstream
It is not dying, it is not dying.Lay down all thoughts, surrender to the void
It is shining, it is shining.
I’d sing the first lines to myself on every descent, telling myself to take the opportunity to switch off my mind, to completely relax, and to enjoy whizzing down the hill in the dark at 60km/h.
Pretty idyllic. What could possibly go wrong? I’d mentioned to James that one possibility was that a rabbit or fox might run out on the road in front of us. I hadn’t thought about badgers though. I saw two of them. One crossed the road running diagonally across and away from me. The second, which was larger, went straight across in front of me at a distance of about 10m. At 60km/h you cover 10 meters in 0.6 seconds. Human reaction time is about 0.5 seconds, so at that kind of distance you basically have no time to do anything. I’d never seen a badger in the wild, at least not alive, and didn’t realize how large they are. If you hit one on a bike at that speed you’d go flying, for sure. Fortunately, nothing else sprung out of the thick bushes and trees on either side of the road, apart from a rabbit or two.
I had resolved to ignore James, apart from not colliding with him when we crossed. I didn’t want to try to match his speed, because I wanted to do the ascents as slowly as I could. At the start I found myself going over 300W sometimes (very easy to do when you’re aiming for 280W) as I watched him ahead of me. He was standing up on the pedals on parts of all his ascents, and generally being James. He was doing laps 10% faster than I was for the first hours. He’d also done 300m more climbing on his way to the start, so our total ascent numbers were off.
After a while I just told myself to watch the power meter for the whole of the ascent. I knew I’d managed to block everything else out when I saw James at some point and thought to myself (half joking, but with a smidgin of truth) “Hell, there’s that same cyclist again. Who is that guy?”
At about 10% into the ride I remember thinking that it felt like I’d done nothing, and thinking “and that’s exactly how it should feel. Good.”
Eventually it began to slowly get light. That gave me an opportunity to snap a photo of Derek in his car “monitoring my nutrition” and “saving his light for me in case I needed it”. Here he is, at 6:40am:
By then, James and I had been riding for 5.5 hours. We’d stopped several times for short breaks (of I guess 5-10 minutes) to eat. We’d open the car and talk to one another, and Derek would sleep right through it. Probably a good thing, as he’d driven to Liverpool and back that day and was going to need his energy.
If you’re curious, I was wearing a Stolen Goat Orkaan Race Tech Weatherproof SS Jersey and their arm warmers. The Stolen Goat jersey is a tech marvel. See this review and watch this video. The first time I rode in the top I was out riding fast (and hence sweating) for 3.5 hours in the rain and it was bone dry inside. There’s a trade-off with the best modern cycling tops – they’re not completely waterproof, in exchange for fantastic breathability, and you still get warmth. I rode in that top and arm warmers all last winter, in the rain (often) and snow (occasional). I also have the waterproof cap shown in the video, and had it in my bag. Amazingly, Stolen Goat is a one-person company, run by Tim Bland.
James was better prepared, as he also had a gilet on. Although the temperature was fine, what I hadn’t taken into account was that the descents were cold. Even though the temperature was 12C at its coldest, going through that at 60km/h is chilly. So I was a little cold. After a few hours I swapped my socks for a warmer pair, the DeFeet Woolie Boolie (currently 1/2 price). It probably seems silly to read about cycling socks if you’re not a cyclist living in a relatively cold country, cycling through the winter, but socks are important. Suffice to say that the Woolie Boolie are fantastic – thick, warm, and breathable.
Garmin’s poor altitude attitude
I had my Garmin set up to show 3-second power average, total elevation gained, heart rate, and lap count. There were plenty of other things I could have displayed (in particular, cadence and time), but I knew what my cadence would be if I kept to my power aim, and I didn’t care about the time. I’d press the lap button at the top of each ascent, and would look at the lap times (typically 3 mins, 30-40 seconds).
Everest is 8,848m and I wanted to see (and photograph) my Garmin go through that total. It seemed a hell of a long way off, but I knew that once I was half way it would start feeling really close and that I just had to keep going. So I was most interested in the total ascent number.
But there was a problem, or a potential problem. I was surprised to note that my lap count multiplied by 36m was higher than the figure the Garmin was showing. I changed the Garmin to show the current elevation and started watching the altitude at the bottom and top of the hill. I was alarmed to see that I was getting about 31m of ascent credited on each lap, and sometimes as low as 29m! Holy shit, I was being ripped off about 15% of my elevation gain by the crappy Garmin barometric pressure sensor and/or calculation. Was I going to have to ride 15% more (over 10,000m) to get the displayed total ascent up to 8,848m?
After a while I found that if I spent a few extra seconds at the top and bottom of each lap that the Garmin would catch up and credit me with the full 36m. So, supposing I added 10 seconds to each lap, that would be 2,500 seconds (41 mins) of time waiting, which was much less time than it would take me to ride an extra 1,300 vertical meters, so I slowed it down.
100 laps in
Our target was 247 laps (though I mainly wanted to hit the altitude number, as well as the lap count). At about 100 laps we were about 40% done. At around that time I found Derek looking fresh, stretching, and smiling on the side of the road at the top of the hill. With his electric toothbrush.
At 1am just before starting, Derek had asked me if I’d checked my tire pressure. I wanted to get started, so I said “yes” and didn’t finish the sentence, which would have ended with “about a week ago”. I’d actually only given them a quick squeeze and thought “she’ll be right.” Near the end of the 100 laps, it was obvious that the rear tire was going to need some air. Looking down and back, and comparing it to the front tire, I could see it was a bit flat. I asked Derek if he could put a bit of air in it and to tell me what its pressure had been. 50 lb per square inch! That’s about half of what I usually put in. He asked if he should also put some in the front and I said I thought it was fine. He did it anyway, and reported that it had been at 60! What a novice error. I’d done 100 laps with greatly under-inflated tires. The ride was nice and smooth, though.
If you’re curious, the received wisdom (narrow, hard) on bike tires is now being repeatedly challenged in carefully conducted experiments on rolling resistance. It’s no coincidence that I was running 28mm Continental Grand Prix 4 Seasons tires.
With the tires both at 100 lbs psi, I found I could get up the hill at 10W less than before. A belated discovery, but nice.
A little before stopping at 100 laps, I’d heard James make a noise as I began the descent and he was just getting to the top of the steepest part of the climb. I wondered what it was and if he’d been trying to say something. A couple of seconds later I realized it had been a grunt of pain. I figured it couldn’t possibly be his legs, but didn’t think too much about it. When we stopped he told me his shoulder and neck were bothering him. He didn’t sound too worried, but I thought that sounded bad, given that we had to keep climbing for at least another 9 hours and that I figured James wasn’t the type to complain, given the kind of punishment levels he’s clearly able to deal with.
Here we are, at about that time, having a break on a bench, with a bit of Derek’s finger thrown in for good measure:
At about 4,000m James told me his wife and kids were arriving and soon after I saw them sitting together at the top. I stopped to say hi and James said he was done. He’d done a 500km ride into stormy weather and a strong headwind a couple of weeks ago, and had ridden several hundred km already last week before the Everesting. I asked him about his gearing. He had a 54/39 elliptical chainring, but said the small ring is actually like a 42 tooth at its widest! And on the back, an 11-28. So his gearing was much tougher than mine. I seriously doubt I could have gotten up the hill more than a few times on his bike. I wondered if it would even be possible for someone like me to ride that bike up that hill without having to stand up on the pedals.
Cue Jeremy
At about 10am, a cyclist with a friendly face had appeared on my left as I reached the top. I knew immediately who it must be, even though we’ve only cycled together once and I couldn’t remember his face. You’re more likely to remember another cyclist’s legs or bike. Only one other cyclist (apart from people I know well and who had declined to join) knew we were making the attempt, and indeed it was Jeremy. He’d been reading and commenting along to the 60+ messages James and I had exchanged in planning the attempt.
Jeremy had obviously chosen his outfit and bike with care. I thought we made a fetching, well-matched team:
The only thing missing was some “Team Mamil” branding for our jerseys.
Unfortunately, his gearing left something to be desired! He had an 11-21 cassette, and given that I don’t think it really mattered whether he had a compact chainring or not (I should think not). He rode a few laps with me, and how he managed to get up the hill I cannot say (I suspected mechanical doping, but was too polite to bring it up). Half way up one ascent I told him that turning up to an Everesting with an 11-21 cassette was like bringing a knife to a pistol fight. I think his reply was mainly a grunt.
Jeremy did a 55+km round trip to come see two guys, one of which he’d only met once before and one he’d never met. How nice is that? Here are his rides, out and back (note the photos), showing his Staunch Hill climbs at the end and start respectively. Jeremy hung out at the top with Derek, and then with James, for an hour and was generally full of good cheer and self-deprecating humor.
Rain!
With James out and Jeremy gone, Derek and I were left alone and I was sitting on about 4,000m. Before he left, James gave me an update on the weather forecast – that rain was coming, as we’d known, but that it was looking ok and should only rain for an hour or so. The first few drops were falling around the time he left.
And rain it did. It was never absolutely pouring, but there was quite a lot of it. Enough to drench me. Hard enough to painfully sting my forehead coming out of the tree cover at 50 or 60km/h on the downhill. Enough to absolutely soak my socks and weatherproof (not waterproof, see above) jacket.
I told myself to just enjoy the rain, not to think of it as a bad thing, that it’s just water, and that many other people had been through much, much worse. It was quite warm, but not so much on the descents in the wind.
And for some added Everesting-in-the-rain cheer, I suggest the theme song from Rawhide
Rollin’, rollin’, rollin’
Though the streams are swollen
Keep them doggies rollin’, rawhideThrough rain an’ wind an’ weather
Hellbent for leather
Wishin’ my gal was by my sideAll the things I’m missin’
Good vittles, love an’ kissin’
Are waitin’ at the end of my ride
performed unforgettably in The Blues Brothers:
After a while I decided to abandon the wet jacket. I dried myself with a towel (Derek thought of everything), and put on a cotton t-shirt and a pretty ineffectual Altura rain jacket that is actually Ana’s (I didn’t think of everything). I knew the t-shirt would soon be soaked, but at least for now I had something dry on. Like an idiot, I had looked at my fantastic fully waterproof Showers Pass jacket that morning, and decided it would be overkill. So I’m out there drenched and about to be re-drenched, and my fine jacket is back at home, dry as a bone, hanging on a hook inside the back door. And probably feeling confused and rejected.
I just ignored it and kept on. I had put my Stolen Goat waterproof cap in my backpack though, and I happily put that on. A warm and dry head, at least.
At maybe 2pm, Derek told me that the weather forecast had changed for the worst. It was now due to rain until 11pm! We estimated that I might be finished at about 9:30pm. The rest of the ride was going to be wet.
Derek’s car battery
Derek drove me to the Tour Of Cambridgeshire race back in June (here’s my ride, which was a total blast). While the race was on, he was in his car and managed to run its battery down. After the race I returned to find the car gone, and cycled around a bit to find him and hear what had happened. Fortunately he’d had his bike with him too, and had been going to ride to a garage to buy some jumper leads, before he happened to meet someone at the race who had some.
So I thought he was kidding when he told me at 3pm or so that he’d run the car battery down and was going to go off on his bike to buy some jumper leads from a garage in Spaldwick. But it was true! He left me the car key so I could get in for food and headed off.
About 10 minutes later I see him heading back along the village road from the church, and he’s no longer alone.
Cue Beth
Beth Evans had told us during the week that she’d cycle to Leighton Bromswold to join in some of the Everesting. She’s a British Military Fitness enthusiast and recently cycled 1500km alone in Eastern europe on an old hybrid bicycle. I asked her if she knew the way, and she said “kind of” in a very untroubled way. I asked how she normally navigated on her bike. “Well, I just go in what I think is the right direction and when I get lost I look on my phone.” And that’s exactly what she did, or I guess tried to do, after finding part of the busway from Cambridge was closed for repairs and following her nose. She’d somehow converted the 50km ride from Cambridge into an 85km ride, in the rain, about which she seemed perfectly happy.
She was tired, and Derek was heading off to the garage, so we suggested she hop in the back of Derek’s car and have a sleep. Did I mention that he had a mattress, two pillows, and a nice-looking comforter in there? So Beth had a sleep and I cycled on.
Later, from the comfort and warmth of the back of the car, she extended just the tip of one finger from the window, and, with perfect deadpan, said “Oh! It’s raining out there.” My kind of humor, I thought, resisting the urge to strangle her.
Much later, with Derek and Beth both warm and dry in the car, I couldn’t resist (well, I didn’t actually try) singing some snippets of Don’t Stand So Close To Me to them:
Young teacher, the subject
Of schoolgirl fantasy
…
This girl is half his age
…
Sometimes it’s not so easy
To be the teacher’s pet
…
Temptation, frustration
So bad it makes him cry
Wet bus stop, she’s waiting
His car is warm and dry
…
It’s no use, he sees her
He starts to shake and cough
Just like the old man in
That book by Nabokov
and then leaving them with an explicit wish that they enjoy some awkward silences.
Ana, Lucas & Findus arrive
Ana arrived with our two boys. So by this time we had 5 people from our Cambridge research group assembled at Leighton Bromswold. Here I am with Ana and Lucas, and then eating a pork pie with Ana and Findus:
After Ana took the boys home she made herself a coffee. Then she thought I might like one and filled a thermos and came back. By that time it was raining and the coffee was very welcome. It’s about a 30 minute drive each way from where we live. So that’s nearly 2 hours of driving to come help twice.
More Garmin woes
At about 5,300m I was having a break and talking to Derek. I went to the bike to check the ascent total and was very surprised to see that my Garmin had returned to display its home screen!
It never shows that screen when you’re on a ride, only when you’re not on one. WTF? “It’s lost the ride” I told Derek. I’ve had so many Garmin crashes and lost so many rides or partial rides that I’ve gotten used to it, and even come to expect it. We looked in the ride history, and indeed the ride wasn’t listed. I tried riding a bit, hoping the screen would change to show the hopefully-still-in-progress ride, but it didn’t. We’d taken a picture of the screen at 5,001m, following the advice of the Hell’s 500 rules page:
Yes, that’s a Garmin 820. About which I should probably write a review.
How had this happened? I’m pretty sure it was the rain. A couple of times earlier in the ride I’d notice my Garmin screen changed. Twice it was on the screen that allows you to change what’s displayed in a data field (so I’d just press the Back button to revert to the original selection). Once I even saw the screen get changed due to a rain drop. The screen is too sensitive. I can activate the touch screen without actually touching it, and very often it mistakes a press for a swipe and does a micro scroll instead of taking you down into a sub-menu. Garmin will hopefully sort all that out with a firmware update. I tried to turn on the screen lock but Garmin have moved that functionality: on the 820 (as I now know) you press and hold the power button, which I would never have tried because on the 810 that turns the unit off.
It was obviously a pain to have the Garmin go on the blink, but not the end of the world. I knew I was at about 5,300m and 160 laps. So we attached Derek’s Garmin (810) to my stem and I set off with both of them, feeling like a pro.
Before starting the descent, I had what seemed like a risky idea. If I pushed the button to stop the ride, and if it happened to still be recording, it might ask me if I was sure (as it usually does) and I could then just hit the same button again to continue the ride. So I went under a tree to be out of the rain (dreading a spurious button press in the wrong place at the wrong time from a raindrop) and tried it. It worked!
But then, just a couple of laps later, another Garmin oddity. I swiped left or right to see another screen, and got this:
And now the unit seemed frozen. It didn’t respond to any screen swiping and when I pressed any button it gave a single beep but otherwise didn’t react. I decided to ignore it and go for a ride. We’d been spending so much time trying to make the damned thing work that I just wanted to get on with riding. Did I mention that I was wet and it was raining?
Then, maybe 10 minutes later, I noticed the Garmin come back to life. In the bottom area of the screen above, it had the beginning of a list of my lap times! So I guess it had been thinking that whole time, and that once it goes into that mode of calculating all that information you can’t interrupt it. I’d actually seen that happen once before, though on a much lower lap count, somehow getting onto that screen due to raindrops, I assume.
While the Garmin had been having its nap, I’d done a couple of laps, and now wasn’t sure if the presses on the lap button would have registered.
A niggling pain
At some point, maybe at about 180 laps, I began to notice occasional short sharp pains behind my left knee. I had no idea what it was, but I still had at least 7 hours of riding to get through. I knew I could make it, as long as I didn’t have a bike problem, an accident, or some kind of injury like James and his neck/shoulder. The knee pain wasn’t bad, but the fact that it was happening was. How would it develop? Would it get worse and worse and force me to abandon as so many others had in the blogs I’d read? Or could it be kept under control? I watched the power meter carefully and saw that it only happened when I got above 210W on the ascents.
So I slowed myself down further, and found that I could take things even easier than I’d thought, and get up the hill without going much over 250W. I’m pretty sure it helped that the wind was up, and helping me with the ascents. My laps were up to about 4 minutes now (up from 3:40), so by slowing down I was adding about 8% to how long it would take me to finish, or about 30 minutes to the 7 hours it would still have taken me to finish (I love doing mental arithmetic on the bike). That was a far better prospect than not finishing at all. On each ascent I’d have a bit of knee pain, and then a chance to stretch the leg at the top and on the descent. I dropped the power on the flat bit at the bottom as well.
It was going to be a long 7 hours though. It was still raining, I was going slowly, and with the knee pain I was wondering for the first time if I was going to make it.
Deus ex machina
Earlier in the day an old guy (anyone who looks older than me is automatically and definitively “old”) had ridden down Staunch Hill and turned around at the bottom. I was doing my gentle ascent as he drew up beside me on his way back up. I said hello, and he replied “This is one of those hills you just do simply because it’s there.” I said “yes, that’s exactly right” and resisted the urge to bring up my tenuous connection to George Mallory. And oh, sweet irony, the guy did a single ascent and breezily rode off to the right when he got back to the top.
So at about 4:30pm, with about 50 laps to go, I was not in the slightest bit impressed to see a flimsily clad young guy zooming down the hill on a bike as I pushed slowly upwards.
And then as he got closer, I realized….. it was DAVID! He had a happy face with a huge smile. He stopped, clapped me on the back, then turned right around and joined the climb.
It was a deus ex machina moment. The arrival of the young Apollo. A powerful boyish god appears in the rain clouds above you, and swoops down into the action to save the day. It’s true that I may have been partly delirious, but it felt miraculous to have him arrive at exactly that moment, also wet, after a 50km ride in the rain from Cambridge.
Whereas Jeremy had been ultra careful and respectful of my riding space at a time when I was quite fresh, David was in my face immediately at a time when I was quite tired. On every single ascent when I moved to the 32 tooth rear cog, I’d said out loud to myself “relax”. I never said that again on the rest of the ride. I think I only thought about my knee pain once in the next 5 hours. Ten minutes after joining, David swerved suddenly and nearly forced me off the side of the road into the bushes during a climb! He rode on the left as we ascended, and I was stuck in the middle having to think about cars, which I hadn’t done at all in the previous 15 hours. He talked non-stop during the ascents.
And it was great, all super super great. I was out of the low spot and back to feeling carefree, happy, and super confident.
And so we climbed, and turned, and climbed and turned. Which reminds me: Brian Eno’s beautiful song, Taking Tiger Mountain had also been playing in my head.
We climbed and we climbed
Oh how we climbed
Climb how we climbed
Over the stars, to top Tiger Mountain
Forcing the lines to the snow
The last I’d heard from David about the Everesting was when he couldn’t make August 27th and James and I had moved the date to the 3rd. David joked that he thought he might have to be out of town that weekend too. After that it hadn’t occurred to me that he might show up. But he and Derek had been talking, and agreed that a good time to appear would be at the end. I’m glad they didn’t tell me, because I’d never have had such a surprise and certainly would never have compared David to a young god :-)
On one of the climbs, I asked him how the climbing pace was for him. He said it was fine, but that it would have been better if he hadn’t given blood two days earlier. Blood is kind of important when cycling up hills for hours on end. So there’s David, doing a 100km round trip just to get to Staunch Hill, in the rain, after donating blood. He was also completely soaked, and had also left his good waterproof jacket at home.
Here he is:
At some point I realized that Derek was sitting in his car in a dry rain jacket (another crappy Altura, but DRY), so it seemed a good moment for a carjacking. He gave me his cycling top and the jacket, so I was dry again, at least for the moment. To my surprise, David said he’d take my soaking jacket and use it instead of the soaking skimpy top he had on. So he put on my wet jacket and declared himself “so much warmer”.
We made a fine pair.
David rode the last 50 laps with me, and we ended with a joint sprint up the final ascent and a big hug. Thanks!
The Garmin decides overall ascent isn’t that important
Soon after David joined, the overall ascent figure on the Garmin simply stopped updating. Great. I remembered it had been on 6,300m and when I looked some laps later the number hadn’t changed. I said something along the lines of “The Garmin never misses an opportunity to fuck you in the arse.”
And there it was, stuck on 6,300m. The other Garmin wasn’t doing too well either. After an encouraging start, where it had awarded me 140m for 4 laps (almost bang on the money), it too had decided that overall ascent wasn’t really worth recording, and had registered a total of just 363m after 40 laps.
My already low opinion of Garmin bike computers had reached new depths. I told Derek and David that disgruntled Garmin employees had probably programmed it to detect when you’re doing an Everesting and to screw with you.
Riding to the magic ascent number was now completely out of the question. We’d have to just do it on lap count. I thought the total laps needed was 257, but Derek checked online and it was only 247!
Just 20 to go.
Derek enters the fray
Derek has a Giant TCR, and it’s not exactly set up for hill climbing (at least not by a regular human) with an 11-25 cassette. He’s also not been riding much this year due to foot surgery. But he joined David and me, pushing himself up that hill over and over, doing much more work than we were. And loving it.
Beth joined us for some laps and then took this photo of the three of us reaching the top of the hill:
Eventually the rain stopped, though the road was still wet and there were puddles at the bottom. The sky took on a wonderful post-storm golden color, the air had that feel of being perfect for photographs, and then things went pink and purple and finally it got dark.
Finishing it
We counted down the final laps. It was totally dark, after 9pm. The rain had stopped. We climbed side-by-side and got to what I guessed was 247 laps. I thought I had missed one lap button press when Jeremy arrived, and I had missed 2 while the Garmin was doing its thing calculating all my lap split times (one of which I had reclaimed mid-lap). So I thought I was off by one or two presses. We decided to do 250 laps, end the ride, and upload it to Strava to see what it made of it.
We did a sprint up most of the final ascent. Probably that was a bit silly because I could have injured that leg properly, but it was ok. There were hugs at the top, and I heard Derek say “Great” about 10 times in a row.
I finally got the Garmin to load the ride, turned on Bluetooth, got it to load the ride again, and uploaded it. Shit! Strava gave it as only 7,000m! Was I going to have to get back on the bike and do another 1800m of climbing, another 3 hours (when fresh), another 50 laps? James had done half the ride and Strava gave him 5,000m and I’d done the whole thing and been given just 7,000m. How was that possible? But at least the lap count was right, 250 of the suckers. At that point I just had to assume Strava would be able to correct the elevation (as it does reliably) and that the Hell’s 500 folks would accept the ride based on the laps and the fact that the hill rises 36m. And so it proved.
The elapsed time on the ride was 20 hours 44 mins, with 16 hours 12 mins in motion. So I had about 4.5 hours off the bike to eat food, socialize, and wrestle with the Garmin. I’d thought I might be able to get done in less time (with 15 hours riding) and maybe 3-4 hours of breaks, but it turned out pretty close. The Everesting calculator had said I’d do 279km, but it turned out to be 315km, so I did an extra 36km over the 250 laps. That didn’t matter much, as it was probably all at the bottom on the flat where it would have been work to stop sooner.
Here’s the ride on Strava, with a screenshot of part of the Strava analysis page for good measure:
Dinner at 11pm
David rode the 50km back to Cambridge with about 5 partially-charged bike lights, despite being asked to stop on the way, come to dinner, and get a ride the rest of the way with Derek. Derek and I got back to the house in Over at 11pm to find the kitchen table set, with candles, wine, champagne, and a ton of food ready.
We ate and then listened to music. Findus asked me if I was ever going to do a ride like that again. I said no, and that there wasn’t really much point – to howls of laughter. They all told me they could have told me that in advance.
After dinner I sent mail to Andy at Hell’s 500 and submitted my ride. Today he replied to say it all looks great, and that I’m now listed in The Hall of Fame and on a special page at VeloViewer. Good.
The fine folk of Leighton Bromswold
We met probably a dozen people from Leighton Bromswold, which is a significant proportion of the total population. They were incredibly friendly, curious, supportive, and kind. Word got around about what we were doing. People pulled over in the cars to ask how it was going and offer encouragement. A vet rugby player (Paul?) offered to make me a sandwich. A woman with a dog asked us if there was anything we needed. Neighbors paused in discussions to say hi and wave. At least one kid on a bike went back and forth many times along the street at the top of the hill, doing his own horizontal flavor of Everesting, shyly looking at us as he passed. It was such a nice environment to find oneself in, totally unexpected, and so friendly.
I’d worried earlier that we might annoy people, turning up in their village at 1am with bikes and bright headlights, doing semicircles at the top of the hill every few minutes, car doors opening and closing, talking, and just generally acting in a very odd way in the middle of the night. I asked a man from the Green Man if they served lunch, and he said yes. But when the time came, I didn’t want to get off the bike for more than 10 or 15 minutes. I had planned to have a beer in the pub at the end, but I was exhausted and cold and dripping wet, and I’d have been drinking alone as Derek had to drive. So the Leighton Bromswold interactions were all cameos. Perhaps that was for the best, and part of the magic.
Short segment Everesting
I think my Everesting was about as easy as they can be. There’s probably no such thing as an easy Everesting – I was cycling for 16 hours in the 21 hours we were out there, 1am to 10pm – but it could have been so much harder. I think the main thing that made it relatively easy was that I had the right gearing (for me). I couldn’t have done it, I don’t think, without the compact and the 11×32. David asked me (twice) if I’d have used an even lower gear if I’d had one, and I said no. The 32 on the back was really perfect. About 1/3rd of the ride in, I made a little joke to Derek and James, casually saying “I feel like I could do this all day”. But that’s exactly how I did feel, like I could go on and on, at this relatively low demand level, staying well within myself. That’s how it should be, of course, because you do have to do it all day. And all night. If getting up the hill in the gears you’ve got hurts you, it’s going to be very very hard to do it for 15 hours.
I’d had two small exploratory rides, the 10% and 20% mentioned above. Those had both pointed at a 15 hour Everesting, assuming I was right that I could keep it going and that things wouldn’t go non-linear. I slowed down deliberately for the knee soreness, and ended up doing ascents with David and Derek at almost exactly the power I’d been using 21 hours earlier at 1am (270-280W). So the low gears made it easy, made it possible to do it almost indefinitely (pretty much without feeling tired, unless I stopped and got cold and stiff, which I soon learned to avoid), and made the overall ride dynamics linear.
I think short segment Everesting is probably much easier (and probably more boring, though I was never bored) than if you do it on a long segment. On a short segment you can have friends hanging out at the top and see them every few minutes. You don’t have to carry anything: no bag, no bike tool kit, not even a water bottle. On a short segment you can much more easily pick a nice piece of road. Cars don’t go as quickly. You don’t spend a long time descending and getting really cold in the wind. And physiologically I’m much more suited to doing short segments with little rests. I’ve always been that way, whether in training for rugby, or athletics, or speed skating. I’d much rather push for two minutes and relax for one and do that hundreds of times than push for an hour, relax for 20 minutes, and do that 15 times.
I’m not sure I’ll ever do another Everesting. Today, the day after, I feel pretty good. Some leg stiffness of course. The back of the knee has some slight muscle or tendon strain that I could feel when I got up, but no longer. I can walk up and down stairs almost normally. So I feel like I got off very lightly. As the foul-mouthed and tremendously entertaining durianrider repeatedly emphasizes: have the right gears and spin up the hills at a high cadence (I was going up the steepest part at 85-95 for most of the day and about 75-85 later on).
Keeping things in perspective
Finally, I found it helpful to keep things in perspective. My Everesting was pretty easy. I got to pick the route, and it was a great one, and I didn’t carry a thing. It rained a bit, but the temperature was fine and I was never a few minutes walk from a car, food, and friends. I had the right gears and I was never climbing for more than a couple of minutes.
Although I find the Garmins endlessly annoying, by far the overwhelming feeling is that they’re incredible. Compare using an old paper map to having a color screen Garmin that has a live map with directions, measurement of everything under the sun, connected to phone, GPS, internet (via wifi), heart meter, power meter, cadence meter, and speed meter.
Compare my easy Everesting with what people go through on the Transcontinental Race (TCR), or the Tour Divide, just for example. Mike Hall, who organizes the TCR, won the 4,400km Tour Divide this year in 14 days (that’s 315km per day – right on my Everesting distance), and the ride has the equivalent of nine Everestings in it, not to mention many other inconveniences, including bears.
And even that’s not so bad, in a way. You’re not living in a war zone, or starving to death, or imprisoned or being raped or tortured. Riding up and down a small hill on an expensive carbon bike in waterproof clothes with a bunch of food and friends on hand in a friendly English village doesn’t feel like much, really. So many humans endure so much more. Some on a daily basis.
To me it was helpful to keep all that in mind. It’s just a bike ride. Keep your hat on.
Trump After the Inauguration
23:17 June 13th, 2016 by terry. Posted under me. | Comments Off on Trump After the Inauguration
Try listening to William S. Burroughs reading his Roosevelt After the Inauguration piece and everywhere you hear “Roosevelt”, think “Trump”.
Burroughs is probably highly NSFW, but Trump is even less suitable for the presidency.
Knog Milkman bike combination lock security flaw
13:51 June 4th, 2016 by terry. Posted under me. | Comments Off on Knog Milkman bike combination lock security flaw
[Note: This issue has been fixed by Knog – see final paragraph.]
I have a Knog Milkman combination lock for my bike. It’s not supposed to be high security – it could be cut through very easily compared to a robust D lock. It’s for when you leave your bike outside a cafe and you’re inside, just a few seconds away, keeping an eye on it. The idea is to prevent opportunistic bike theft.
I like the design and manufacture of the lock, but have realized there’s a security flaw.
The numbers on the lock are just painted on and, unfortunately, the paint comes off very easily. I’ve taken it on rides maybe 10 times. I carry the lock in a rear jersey pocket, perhaps with a gel or a powerbar, but never with anything you’d consider abrasive. You can see the state of my lock in the photos above.
At first I thought the paint coming off was just an inconvenience, but then I realized it will also (typically) greatly reduce the lock’s security.
The problem is that after unlocking your bike you’re most likely to put the lock away as is. Because the position on the lock where you line up the correct numbers to unlock is at the lock’s widest point, it’s that row of numbers that gets its paint rubbed off fastest.
If you look at the above front and back photos of my lock, you can see what I mean. The numbers 8578 on one side have been completely obliterated and (adding 5 to each number) 3023 are greatly degraded on the other.
Because I always just put my lock straight back into my jersey pocket, that means that my combination is being rubbed off! It means that anyone wanting to open my lock would only have to look at 16 different combinations (3 or 8 at the first position, 0 or 5 at the second, 2 or 7 at the third, and 3 or 8 at the last). Sixteen combinations to try is a lot less than 10,000.
You can mitigate this situation by giving your lock a random twist before putting it in your jersey pocket. But then over time all the numbers will have their paint rubbed off. Or you could always set your lock to 0000 after unlocking, so just 0000 and 5555 would get rubbed off.
Of course the best solution would be for Knog to improve the numbering so it doesn’t come off in the first place. I sent them a mail last week (via the form on their web site) to tell them of this issue, but they’ve not responded.
Update (June 22, 2016): Knog responded a day or two after I wrote the above. They asked for photos of the lock, and I pointed them to this page. I was a bit worried there might be a knee-jerk upset reaction because I’d posted images of the problem here, but there was nothing of the sort. Instead, I’m told the numbering issue has been fixed and they asked for my address to send me a new lock :-) Given how often companies react badly to things like this (even though this is a very minor issue, seeing as the lock isn’t supposed to be high security in the first place), it’s very nice to see a mature non-hysterical (or even legal) response. Thanks, Knog!
I go shopping for a compass, then my Sonos decides it needs one too
17:12 December 10th, 2014 by terry. Posted under companies, other, tech. | 1 Comment »
 Last night I spent some time online looking to buy a compass. I looked at many of the Suunto models. Also yesterday I installed Little Snitch after noticing that an unknown
Last night I spent some time online looking to buy a compass. I looked at many of the Suunto models. Also yesterday I installed Little Snitch after noticing that an unknown gamed process wanted to establish a TCP/IP connection.
Anyway… a few minutes ago, 10 or 11 hours after I eventually bought a compass, a message pops up from Little Snitch telling me that the Mac OS X desktop Sonos app was trying to open a connection to ns.suunto.com. See image on right (click for the full-sized version).
WTF?!?!
Can someone explain how that works? Either Little Snitch is mightily confused or…… Or what? Has the Sonos app has been digging around in my Chrome browser history or cache or cookie jar? Is Chrome somehow complicit locally? Or something with cookies (but that would require the Sonos to be accessing and sending cookies stored by Chrome). Or…. what?
And, why ns.suunto.com? There’s an HTTP server there, but its / resource is not very informative:
$ curl -S -v http://ns.suunto.com * Rebuilt URL to: http://ns.suunto.com/ * Hostname was NOT found in DNS cache * Trying 23.63.99.202... * Connected to ns.suunto.com (23.63.99.202) port 80 (#0) > GET / HTTP/1.1 > User-Agent: curl/7.37.1 > Host: ns.suunto.com > Accept: */* > < HTTP/1.1 404 Not Found * Server Apache is not blacklisted < Server: Apache < Content-Type: text/html; charset=iso-8859-1 < Date: Wed, 10 Dec 2014 16:02:47 GMT < Content-Length: 16 < Connection: keep-alive < * Connection #0 to host ns.suunto.com left intact File not found."⏎
Unfortunately, Little Snitch doesn't tell me the full URL that the Sonos app was trying to access.
Anyone care to speculate what's going on here?
Learning jQuery Deferreds published
22:24 January 7th, 2014 by terry. Posted under deferreds, javascript, me, programming, python, twisted. | Comments Off on Learning jQuery Deferreds published
 Nicholas Tollervey and I have written a book, Learning jQuery Deferreds, published by O’Reilly.
Nicholas Tollervey and I have written a book, Learning jQuery Deferreds, published by O’Reilly.
If you’ve been a reader of this blog over the years, you may have noticed that I’m very fond of deferreds (also known as promises or futures). I’ve mainly been using them in Python with Twisted, and a couple of years ago was happy to notice that jQuery also has a (somewhat different) version of deferreds. Asking around, it soon became clear that although there are tens of millions of programmers who’ve used jQuery, very few of them have ever used deferreds. E.g., at the jQuery conference in San Francisco in 2012, only about 25% of the audience in a talk on deferreds. There are about 19,000 results for “deferred” on StackOverflow.
This seemed like a perfect storm: a fantastically cool and empowering technology that I love thinking and writing about, built in to a ubiquitous web library, in use by millions of programmers, and yet somehow not widely known or used.
The book tries to really teach deferreds. There are 18 real-life examples, along with 75 challenges (and solutions). We’ve tried to get readers into deferreds the way we are, to be provocative, to try and get you thinking and scratching your head. To get you to see how cool and elegant working with deferreds can be. In short, to make you one of us.
Although the book focusses on jQuery’s flavor of deferreds, we wrote it with a much broader aim: to be just as useful to people working with any flavor of deferreds in any language. The concepts behind deferreds are few and simple. Even if you’re not a jQuery user or a JavaScript programmer, we hope you’ll enjoy and benefit from the book.
If you’re curious, the animal on the cover is a musky rat-kangaroo. O’Reilly chose it for us. When I first saw it, I mailed them and Nicholas to say it looked “overfed, passive, and thoughtful” to which Nicholas replied that it resembled me. The O’Reilly toolchain is modern and fun, employing AsciiDoc and a shared Git repo. We wrote 30,817 words and 2,301 lines of Javascript. There’s a source code repo for all the book examples, at https://github.com/jquery-deferreds/code. We spent six months on the book, during which I usually spent 1-2 days a week on it. It was a blast to write.
If you’d like to buy a copy, use AUTHD as a discount code on the O’Reilly site and you’ll receive 40% off the print or 50% off the e-book. Please let me know what you think, and/or leave a review on the O’Reilly (or Amazon) site. Have fun, and thanks!
txdlo – a Twisted deferred list observer
21:46 December 30th, 2013 by terry. Posted under deferreds, programming, python, twisted. | Comments Off on txdlo – a Twisted deferred list observer
Last night I wrote txdlo, a Python package that provides a class called DeferredListObserver. As you might guess, it lets you observe callback and errback events from a list of Twisted deferreds. You can add observers that will be passed information about deferreds firing. You can add deferreds to the observed list at any time, which is very useful if you’re dynamically creating deferreds that you want to monitor.
The class can be used to easily build functions or classes that provide deferreds that fire when arbitrary combinations of events from the observed deferreds have occurred.
For example you can write functions or classes that support deferreds that:
- Implement Twisted’s
DeferredListor simple variants of it, or that let you separate the various behaviors of DeferredList into simpler functions. - Provide a deferred that fires when N of the observed deferreds have fired.
- Provide a deferred that ignores errors until one of the observed deferred succeeds, only firing with an error if all the observed deferreds fail.
- Or (a more involved example), suppose you have 3 methods that can return you a user’s avatar: a fast local cache, a filesystem, and a slow network call to Gravatar. You want to write a deferred-returning function that launches all three lookups at once and fires its deferred with the first answer. But if the cache and/or filesystems fails first, you don’t want to trigger an error, you instead want to take the result from Gravatar and add it to the cache and/or filesystem, as well firing the returned deferred with the result (wherever it comes from). Only if all three lookups fail do you want to errback the deferred you returned.
Here’s a sample example usage, which shows how to use DeferredListObserver to build a simplified version of Twisted’s DeferredList class as a function:
from twisted.internet.defer import Deferred, succeed
from txdlo import DeferredListObserver
def deferredList(deferreds):
"""
Return a deferred that fires with a list of (success, result) tuples,
'success' being a boolean.
@param deferreds: a C{list} of deferreds.
@return: a L{twisted.internet.defer.Deferred} that fires as above.
"""
if len(deferreds) == 0:
return succeed([])
dlo = DeferredListObserver(maintainHistory=True)
map(dlo.append, deferreds)
deferred = Deferred()
def observer(*ignore):
if dlo.pendingCount == 0:
# Everything in the list has fired.
resultList = [None] * len(deferreds)
for index, success, value in dlo.history:
resultList[index] = (success, value)
deferred.callback(resultList)
dlo.observe(observer)
return deferred
You can grab the code, read more about usage, and see several other examples at https://github.com/terrycojones/txdlo.
Promises are first-class objects for function calls
01:14 September 12th, 2013 by terry. Posted under deferreds, javascript, programming, python, twisted. | 17 Comments »
 Have you ever programmed in a language without functions as first-class objects? You can’t return a function from a function, can’t pass a function as an argument, and you certainly can’t make anonymous functions on the fly. Remember how liberating, empowering, and flexible it felt when you moved to a language with functions as first-class objects?
Have you ever programmed in a language without functions as first-class objects? You can’t return a function from a function, can’t pass a function as an argument, and you certainly can’t make anonymous functions on the fly. Remember how liberating, empowering, and flexible it felt when you moved to a language with functions as first-class objects?
Yesterday, after 7 years of working with deferreds/promises/futures (call them what you will), thinking carefully about them thousands of times, mailing and blogging about them, giving talks about them, and now writing a book about them, I realized there is a very simple way to look at them:
Promises are first-class objects for function calls.
In other words, a promise gives you a way to make a function call and to pass around that function call. Return it from a function, pass it as an argument, put it in a data structure, etc. Given a promise, you can arrange to process the result of the function call or its error.
To be a bit more abstract: A promise is a time-independent reification of a function call.
By “time-independent” I mean that if you get a promise from somewhere, you don’t have to worry whether the underlying function call has already completed, is currently in progress, or has yet to run. Depending on the promise implementation there may be no way to find out. The point is that you don’t have to care and you shouldn’t care.
That’s all I’ll say for now, except for a final comment on naming.
I think “promise” is a better name than “future” or “deferred” because the latter two feel more like they’re referring to an event yet to happen. A promise feels more neutral: you could easily be making a promise about something you’ve already done. Many explanations of promises/deferreds/futures stress that they are something that will hold the result of a computation that’s not yet completed. That’s certainly a common usage, but it’s only part of the picture. Describing promises as a reification of a function call takes the time factor completely out of the picture.
Here’s a small Javascript function to memoize a (single-argument) function to illustrate the point:
var memoize = function(fn) {
var promises = {};
return function(arg) {
var promise = promises[arg];
if (!promise) {
promise = promises[arg] = $.when(fn(arg)).promise();
}
return promise;
};
};
The memoization cache is full of promises, not underlying function results. Some of the function calls may have completed, some may be underway. I’m using the jQuery $.when function as a convenience (that’s an irrelevant detail, don’t let it distract you). The promises stay in the cache indefinitely (no eviction, for simplicity), holding promises for function calls from the past.
Time is not an issue here.
Specifically, and in contrast, think about what happens with non-promise memoization. What happens if a first call comes in with an argument X, but before fn(X) has finished computing there is another call with argument X? Then another and another and another? You wind up calling fn(X) many times. I.e., doing exactly the thing you were trying to avoid! And there’s nothing you can do about it.
If you’re interested to review the book I’m writing with Nicholas Tollervey on jQuery deferreds, please email me or leave a comment below. Most of the book is not really specific to jQuery’s flavor of deferreds.
The mockery pervading human affairs in all their aspects
23:22 September 5th, 2013 by terry. Posted under books. | Comments Off on The mockery pervading human affairs in all their aspects
 From Tacitus, on the eventual rise of Claudius to Roman emperor, from The Annals of Imperial Rome (Penguin Classics, p127):
From Tacitus, on the eventual rise of Claudius to Roman emperor, from The Annals of Imperial Rome (Penguin Classics, p127):
The more I think about history, ancient or modern, the more ironical all human affairs seem. In public opinion, expectation, and esteem no one appeared a less likely candidate for the throne than the man for whom destiny was secretly reserving it.
The Cambridge Companion to Tacitus (p169) translates as follows:
The more I consider recent events or those in the distant past, the more I am confronted by the mockery pervading human affairs in all their aspects. For in public opinion, expectations and esteem, no one was less likely a candidate for the throne than the man whom fortune was secretly holding in reserve.
1793 viruses!
01:23 July 30th, 2013 by terry. Posted under me. | 4 Comments »
In case you missed it, I spent ten days in hospital this past May (2013).
When they took the skin biopsy from my arm, I got them to take 2 samples. One of them, along with a throat and skin swab was later sent to the virologists I do some work with in the Viroscience Lab at the Erasmus Medical Center (EMC).
I got the sequence data back about a week ago and have been looking at them, firstly via BLAST and then using a bunch of code I’ve been writing lately. There are 115K reads from 6 preparations (RNA and DNA protocols for each of the 3 samples). These come from a “next generation” sequencer, a Roche 454. The next generation sequencing involves using random primers to indiscriminately match genetic material. My BLAST output files are about 82Mb in total (this is relatively small, some of my other data sets are about 30Gb). I BLASTed against a viral subset alias nucleotide database that I made from the full NCBI Nucleotide Database, excluding all bacteriophage viruses. There are about 1.3M viral sequences in the subset db.
I wont go into details, but wanted to dump a bit of data that’s pretty amusing / interesting. Just to give the EMC folks an idea of the scale of diversity I am seeing, I grepped out all “complete genome” hits from all the BLAST output. I chucked out suffixes in the sequence titles that matched the regex (nearly )?complete genome|isolate|strain|subtype).* and then stripped the titles of any text beyond the string “virus” in the title (this step collapses a lot of virus strain information that should really be kept). Then, do a unique sort and…. it turns out I have reads matching at least 1793 viruses.
I feel like the subject of a metagenomics study. At the hospital, once the chickenpox tests had come back negative, they threw a ton of tests at my samples and everything was negative. Otherwise, I’d really be worried :-)
Given the list of sequence matches, it feels like the only plausible explanation is that I’m actually dead and that this is all just a simulation.
jQuery-when2 – a more flexible way to work with jQuery deferreds
21:01 July 27th, 2013 by terry. Posted under deferreds, javascript, programming, tech. | Comments Off on jQuery-when2 – a more flexible way to work with jQuery deferreds
I’ve often been frustrated by the limited functionality of jQuery.when. You give it some deferred objects and it fires when all the deferreds are finished. The trouble is, if any of the deferreds is rejected the deferred returned by jQuery.when fires immediately. So you can’t use it to collect the results of all deferreds including any errors. You also can’t use it to fire when the first of the passed deferreds fires.
So last night I wrote a new version of when, called jQuery-when2 that offers the three behaviors that I commonly want:
- resolve on the first success,
- fail on the first error (the
jQuery.whenbehavior), or - resolve when all results (successes or errors) have been collected.
The API differences from jQuery.when:
when2must be called with a list as its first argument.- An
optionsJavascript object may be passed as a second argument. - If
options.resolveOnFirstSuccessistrue, the deferred returned bywhen2will resolve as soon as any of the passed deferreds resolves. In this case,.donecallbacks will be passedindexandvalueargs, whereindexis the index of the deferred that fired. If non-deferreds are in the arguments towhen2, the first one seen will trigger the resolve (not very useful, but consistent). - If
options.failOnFirstErrorisfalse, the deferred returned bywhen2will never fail. It will collect the results from all the deferreds and pass them all back. The caller gets to figure out which values (if any) are errors. - If no options are given (or
options.failOnFirstErroristrue),failwill be called on the deferred returned bywhen2on the first error (this is the behavior ofjQuery.when). The args passed tofailwill beindexandvalue, indicating which deferred was rejected. - Any
.progresscallbacks added to the returned deferred are also called withindexandvaluearguments so you can tell which deferred made progress.
You can grab the source code, see examples, etc., on Github.
BTW, I’m writing a short (about 75pp) O’Reilly book, Learning jQuery Deferreds, with Nicholas Tollervey. If you’re interested in reviewing early Rough Cuts drafts, please let me know! The book will be out late this year.
Yet another cancelable Twisted deferred class
23:15 June 20th, 2013 by terry. Posted under deferreds, python, twisted. | Comments Off on Yet another cancelable Twisted deferred class
I’m posting this (completely untested!) to illustrate how one could write a class to provide a Twisted deferred-like class, identical to the twisted.internet.defer.Deferred class, but which lets you call callback, errback, or cancel on the instance yourself. Hopefully that will make some sense. If not, let me know in the comments and I’ll try to be clearer. There’s also discussion of this issue (again) in the Twisted mailing list right now. For context, the discussion thread from January 2010 where I first wrote a class like the below is here.
from twisted.internet.defer import CancelledError, Deferred
from twisted.python.failure import Failure
class ControllableDeferred2013(object):
"""A Deferred-like class that takes a regular Twisted Deferred and
provides a deferred that can be fired at will. If you have a regular
Twisted Deferred, you can produce a deferred you have more control over
by using your Deferred instance to make an instance of this class.
Any time you need to fire a ControllableDeferred2013 instance for any
reason, call its callback, errback or cancel method. It will fire
immediately with the value you provide and the original Deferred will
be cancelled."""
def __init__(self, originalDeferred):
self._fired = False
self._originalDeferred = originalDeferred
self._newDeferred = Deferred()
for method in ('addBoth', 'addCallback', 'addCallbacks', 'addErrback',
'chainDeferred'):
setattr(self, method, getattr(self._newDeferred, method))
originalDeferred.addBoth(self._originalFired)
def _originalFired(self, result):
if not self._fired:
self._fired = True
self._originalDeferred.chainDeferred(self._newDeferred)
def cancel(self, value=None):
if not self._fired:
self._fired = True
self._newDeferred.errback(Failure(CancelledError(value)))
self._originalDeferred.cancel()
def callback(self, result=None):
if not self._fired:
self._fired = True
self._newDeferred.callback(result)
self._originalDeferred.cancel()
def errback(self, fail=None):
if not self._fired:
self._fired = True
self._newDeferred.errback(fail)
self._originalDeferred.cancel()
def pause(self):
self._newDeferred.pause()
self._originalDeferred.pause()
def unpause(self):
self._newDeferred.unpause()
self._originalDeferred.unpause()
Ten days in hospital
14:01 May 23rd, 2013 by terry. Posted under me. | 13 Comments »
I’m about to check out of Addenbrookes Hospital in Cambridge after a 10-day stay, 8 of them in isolation. The short story: I got a rash, and it took over my body. Below are some notes on what’s been going on, along with a few images. You can see the full set of images at http://bit.ly/terry-rash (you may need to register for Dropbox).
April 29 – May 5: The LCHF diet begins
 Just back from a week in New York, I decided to start a low carb, high fat (LCHF) diet. I’ve put on about a kilo of weight a year over the last 9 years and eating less carbs seemed like it might be a good way to start burning some of my excess fat. In 2012 I’d done some reading about LCHF diets but hadn’t tried it (check out this video if you’re curious). In NY I’d been feeling too physically large, which prompted me to give it a go. After almost 50 years of living on cereal, bread, pasta, rice and potatoes, though, I wasn’t sure I’d be able to do it.
Just back from a week in New York, I decided to start a low carb, high fat (LCHF) diet. I’ve put on about a kilo of weight a year over the last 9 years and eating less carbs seemed like it might be a good way to start burning some of my excess fat. In 2012 I’d done some reading about LCHF diets but hadn’t tried it (check out this video if you’re curious). In NY I’d been feeling too physically large, which prompted me to give it a go. After almost 50 years of living on cereal, bread, pasta, rice and potatoes, though, I wasn’t sure I’d be able to do it.
But it was suprisingly easy. I’d cook myself some bacon and eggs for lunch, along with a big salad, maybe a fried vegetable, nuts and dried beans for snacks, etc. Within a few days I was clearly in ketosis. I wasn’t getting hungry at all because my body had switched to burning my fat, and there was still plenty of that to consume. After 4 days I was down 3kg (from 78 to 75), or about 6.5 pounds. I felt like I was eating more healthily than ever.
Monday May 6: The rash appears
On the morning on Monday May 6, I awoke with a small itchy rash on my sternum, just small red separated dots, probably about 20 of them. I didn’t think too much of it.
Tuesday/Wednesday May 7/8: Initial spread
The rash grew across part of my chest and down my stomach. On the night of May 8th I slept on a fold-out bed downstairs so as not to bug Ana through the night. The rash was extremely itchy, but I knew I must not scratch it. I lay in bed wishing there were manacles by my sides to hold my arms down.
I had began searching around on the web for LCHF rash connections and found there were many hundreds of people blogging and commenting on forums about their rashes. But I couldn’t find anything that looked like reliable scientific opinion on what was happening to those people. Most of them had small rashes and many were able to stop the rash simply by reintroducing carbs. I added some carbs back to my diet, but there was no change. Almost all the comments online were reassuring, saying it was normal, that (unspecified) “toxins” were leaving my cells and exiting through the skin, etc. As so many people had similar problems, and they seemed to just go away after a while, I wasn’t too worried, just very itchy.
Thursday May 9
I had to take Findus to a doctor appointment after he got back from school. While there I showed part of my stomach to the doctor. She immediately said it was an allergic reaction. We didn’t discuss it further. I’ve never been allergic to any food, and hadn’t been eating anything new – just different amounts of foods I’d always eaten, and very few carbs. The doctor said she highly doubted the rash could be due to the diet change. She suggested I pick up some calamine lotion, hydrocortisone cream, and anti-allergy tablets to see if any of them helped.
By this time, the fourth day, the rash had spread across under my armpits and slightly down my upper inner arms. It was all across my stomach, and my belly button was all rash (see image below). It had begun to descend onto my upper thighs. It was on my lower legs, just above my ankles, and on the back of my calves. My lower back was covered with it. Did I mention that it was itchy as hell?
Friday May 10
I went into Cambridge to meet people I’m doing some part-time research with (on virus detection and discovery, ironically). I was uncomfortable the whole time and spent much of the several hour meeting standing up. Sitting down was causing uncomfortable rubbing pressure on my pants line.
Late that night, Ana convinced me to call the emergency medical line. I didn’t feel it was warranted, but I admit I was worried. Plus, it was Friday night, the local surgery would be closed until Monday, and it was abundantly clear that if this thing didn’t stop spreading it was going to be really bad by then. A woman took my details, listened to the description, and decided to have the medical team call me. They did, and at 11:30pm I was given a midnight appointment at the Chesterton Medical Centre in Cambridge.
The doctor who saw me listened to my story and said I was having an allergic reaction. He put a bunch (8?) of triangular pink steroid tablets into a cup of water and got me to drink it. He wrote me a prescription for more. While talking to him I noticed an unusual slight twinge in my left eye, but didn’t think to say anything.
That night, the rash began to ooze. I noticed when turning over in bed that the sheets seemed to be wet. At first I thought I must be sweating a tiny bit without noticing it. After a while I realized the liquid was coming out of my body in tiny slippery beads of rash pus. Wet areas appeared on the sheets corresponding to my back, stomach, sides, thighs, etc. Very unpleasant. I didn’t sleep well, and not for the last time.
Saturday May 11
The rash has again spread and become denser. I filled the prescription and went into Cambridge with Sofia to meet Ana and the boys. Back home I took my first steroid dose. My right eye had begun to twinge too. Liquid-filled blisters were beginning to appear on the palms of my hands and on my fingers.
Sunday May 12
After another unpleasant night and more rash spreading, I called the emergency medical people again and got an immediate appointment at the Chesterton Medical Centre. I didn’t take calling for help lightly, but I was constantly uncomfortable and the steroids didn’t seem to have helped at all. The opinion this time was chickenpox. The doctor examined my eye but found no lesions on the cornea. So I went off the steroids and onto aciclovir tablets and aciclovir eye ointment. I was told to go to the Over Surgery on Monday to get a blood test for detection of chickenpox antibodies to confirm the dignosis. I mailed my parents to see if they could remember if I’d had chickenpox already. It seemed very likely that I must have, given that I did not get it when our 3 kids did.
Monday May 13: In isolation at Addenbrookes
I’m not saying much about the physical side of things. The rash itself was mainly itchy. But my upper body had become extremely sensitive, and I could hardly bear for things to touch it. The sensitivity had been making me tense for some days, so I was not physically relaxed – far from it. My breathing wasn’t natural and even: I’d take in a breath and hold it a little to avoid breathing out and moving my body. Looking in the mirror at my whole body was quite shocking. Given the speed of the spread of rash and the discomfort level, it was clear that in a couple of days things would be really serious.
I called the Over Surgery at 8:30am and got a 9am appointment to have a blood test. I packed a few things in a small bag because I had a feeling I might not be back for a while. As it turned out, I wouldn’t be back home for 10 days.
On arrival, they sent me to a small room where a nurse did the blood samples. She asked why I wanted the test and I told her. I lifted up my top to show her some of my stomach and she said “I’m calling a doctor”. When the doctor did not arrive after a few minutes, she called another. They both soon arrived, and both said the same thing: go straight to hospital.
It’s about a 25 minute drive to Addenbrookes hospital from Over. They tried calling Addenbrookes to speak to the dermatologist, but couldn’t get through. So the doctor wrote me a letter to help with admissions and told me to go to the Accident & Emergency section. I would have much preferred to go in an ambulance than to drive. I guess uncertainty about driving showed in my face or body. The doctor said they could call one, but I declined. I went out and folded myself uncomfortably into the car. I knew I could hold on and keep it together, but I also felt like I was on the edge of some kind of collapse due to sensory or stress overload. It’s hard to explain, but I didn’t feel well at all. I told myself to focus, to act normal, turned on the radio and off I headed.
At Addenbrookes I drove into the carpark. UK parking places are invariably narrow, and I wasn’t looking forward to trying to get into one. Turning my body or stretching to see were painful. Up and up I crawled in a line of cars looking for places, all the way to the 6th level before finding a spot. Then a short walk, feeling a bit geriatric, to A & E.
Inside, they screen all arrivals. I handed over my introduction letter, was given a summary form, and told to sit and wait to be registered. I looked at the summary form, which had the usual mundane information: name, address, etc. At the top, a description of the reason for the visit. Mine said “Rash. Cause?” I was soon called to the registration desk and the woman took more details, including next of kin, and asked if anyone knew I was there. She was telling me I’d need to go sit down again to wait, but I needed attention. I said: “I know ‘rash’ doesn’t sound like much of a problem, but I think I need help right away”. I showed her my stomach. She took one look and sent me back to the woman doing the input screening, and told me to tell her my story. I showed the screening woman my stomach, and that was it. I was out of the admissions area at high speed.
In a small room, they asked me a few questions. I was already in the medical system as “Doctor Jones”. I knew I now had to mention that I’d been doing some work in the Viroscience laboratory at the Erasmus Medical Centre (EMC) in Rotterdam a few weeks earlier. As I wrote to my father later that night in email:
If you ever want to get expedited at top speed through hospital admissions, being called DOCTOR JONES, having spent recent time in one of the world’s top viral labs, and having an unexplained full-on bright scarlet 90%-coverage body rash ain’t a bad combo!
They took me straight to a small isolation room. When they left me there I heard them hang something on the door on the outside – it sounded like they were barring it! Nurses and doctors began to come by to ask questions, take blood samples and nasal/oral swabs. I had a canula line put into the back of my left hand. My heart rate was 120 on admission. Resting it is 60 or less. A Scottish nurse asked to see my back and exclaimed “Oh, you poor, poor, gentleman!”. I had no idea what my back looked like. Bad, I guess. The questions were about recent travel, what I had been doing in Rotterdam, medication, health history. Chickenpox, excema, other illnesses, allergies, the diet change. Sexual practices, drugs, medication, travel, animal exposure, etc.
About 5 hours after arriving, I was admitted and told I wouldn’t be going home that night. After a chest x-ray, I was transferred to an isolation room in Ward L4. It was a ward full of older post-surgery patients, not one used for infectious disease patients. But they needed an isolation room, and that’s what was available. The room was huge, with a window and en suite bathroom. Just as I arrived, I ran into Ana and Derek in the ward looking for me. I was happy to be in hospital, being looked after instead at home looking at my body with mounting dread.
Derek took some photos and emailed them to medical and virology friends at EMC to get their opinions and to ask if anyone else at EMC had come down with a similar condition. The prevailing thinking, at least at Addenbrookes, was that I had chickenpox. The blisters on my palms seemed the strongest indicator. Rather few diseases cause blisters, apparently. My parents had mailed me back to say they couldn’t be sure whether I’d had them.
Tuesday May 14
The night was long and uncomfortable. I wont go into details of the discomfort (see below for some of that). It finally got light around 4:15am. I spent hours standing around in the room naked, sometimes with a blanket around my shoulders but held off my body. I’d lean against the wall to stretch my back, and listen to the clock tick. At some point I realized the rash had gotten into my scalp. I hope my face will continue to remain free from it.
I don’t remember much about the day. Blood samples, swabs, antibiotics going into my arm, more doctors and more questions. Ana and Derek visited again. Ana brought fruit and other supplies, including my Nexus 7. I had almost no comfortable positions that I could maintain for more than a handful of minutes, but lying on my back was fine. The Nexus 7 would become my point of contact with the outside world. The small screen, a case allowing it to stand propped up on my chest, some of my music on it, and connected via a wifi hotspot made by my phone – perfect. I couldn’t have used a laptop.
I met Dr Sterling (head of Dermatology) that day and was very impressed. I also had a swarm of 5 female dermatologists staring intently at my skin as I stood naked in front of them. I told them I wished I had a camera.
An old woman named Margaret has moved into the room next door. She’s confused and disoriented. Every 5 or 10 minutes she walks out of her room and announces “I’m going home”. The nurses tell her she needs to go back to her room, that she’s in hospital, that she can’t go home. “I’m not a child you know, I’m going home.”
Wednesday May 15
Margaret wanders silently into my room as I’m eating breakfast. I don’t turn around, thinking it is a nurse. Her voice comes from behind me: “wrong house.” I don’t say anything, and she leaves. Later in the day, I am taking a pee and the door to my bathroom is open. I hear someone come in and call out to tell them I’m in the loo. There’s no response. Then I hear a voice call “Margaret, you’re in the wrong room.” Margaret calls back to them “my sister is here, she’s in the toilet.” By the time I come out of the bathroom, Margaret is gone. Later I hear them telling her that she’s going home today: “I certainly am not!” she replies, telling the staff that her friends are coming here to visit her and that she has to leave the ward to find them. No, no, they reassure her, your friends are coming here, they’ll come here to where you are. “But why would they come here?” I feel sorry for them all. Poor Margaret, and the nurses who have to look after her constantly coming out to declare she’s leaving and needing to be shepherded around the ward.
The coalesced parts of rash have gone a dark purple color. Meanwhile the rash has again spread and gotten denser everywhere. Blisters (at least 50 of them) are spreading on my palms and fingers. The VZV (Chickenpox) result came back negative, as did HSV (Herpes Simplex Virus).
With the negative chickenpox result, the case is suddenly more interesting and pressing. I met Dr Moore (head of Infectious Diseases) who comes to see me with Doctor Sterling. Dr Moore stared at me like a hawk, listened to everything very carefully, and then began asking questions. I liked her immediately, she seemed extremely smart and thorough. She suggested they move me to her ward tomorrow so I could be closer to the infectious disease people.
Tomorrow they’ll start throwing every test at me they can think of. Although the rash might be treatable with a steroid cream, the steroids suppress the immune system. So we need to figure out whether my body is busy fighting an actual disease before using steroids to try to settle what could otherwise be some kind of allergic reaction. The doctors here and at EMC start thinking of possibilities while the rest of us are digging for possible leads in Wikipedia, aided by Google Images.
What the hell have I got?
Thursday May 16: Skin pain like on that holiday trip to Mercury
The canula in my left hand has been hurting a lot when they pump in the antibiotics. So they’ve changed it, and put it into my right forearm. Unfortunately, the plastic dongle attachments hang down to the middle of my forearm where it is unbearably sensitive. Shit.
From a mail I sent to Ana & Derek later this night:
Skin pain like on that holiday trip to Mercury, the one where you forget to take any sunscreen.
During the day, Dr Sterling takes two skin biopsies taken from left forearm. Five stitches in them, in total. One of the samples will go into her -80C freezer and we’ll send it to EMC if necessary.
That night I have a pain in my trunk, back right, as I get up out of a chair. Derek is still there and I tell him. After 15 minutes of waiting for it to go away, we alert the staff. I’m almost unable to raise myself in bed to a sitting position so as to get up. One problem they’re watching out for is any kind of internal infection, so a pain in my right kidney region is a bit of a worry. I’m sent for a chest x-ray in wheelchair, and I have to wear a mask. More blood is taken and sent off for septic analysis.
Ahead of the transfer to Infectious Diseases, I have a shower. I have a 500ml container of anti-bacterial soap-like liquid and I’m supposed to wash my whole body. On my left forearm I have the two bandages (covering the stitches from the biopsies) that are not supposed to get wet, and sticking out the middle of my right forearm are two dangling plastic dongles attached to a canula with a tube going into my arm. So I’ve got one arm that can’t get wet and one that I can’t bend properly, and I’m supposed to have a shower and wash myself?
After somehow managing that, I begin putting liquid paraffin all over the rash. I.e., over my entire body. You know you have a real rash when they provide you with soothing ointments by the liter. Once I’m done, the rash feels better, but I am totally covered in shiny paraffin. I hang out naked for as long as I can and then put on the hospital pyjamas for the ward transfer.
I’m transferred at 8pm to Infectious Disease ward D10, and placed in isolation. There’s an air lock to get into the ward and a “Barrier Nursing” sign on my door. The room is about half the size of the one I’d just left, and cold. Derek goes to ask them if they can turn off the blower, but it’s part of the airflow set-up of the isolation ward. He gets me a small radiator to counter the cold and soon after that, around 9pm, they ask him to leave.
I now have about 9 hours to get through before I’ll see the morning staff. I lay in the dark with the Nexus 7 on my chest and sent a short mail to Russell:
This is utterly hellish. The last ward was a paradise. I half expect Derren to walk in. I should be tweeting it. It’s only 2am. I have to look at it as a survival course. I wish I could write more easily. This is left hand only for various reasons. I should make a list.
I didn’t want to expand on how horrible I felt, because it seemed extra words could only make it seem less horrible than it was. But I had nothing better to do, I was certainly not going to fall asleep, so I might as well write up a list of things that were collectively making this all so unpleasant. So from 2-3:20am, typing with the index finger of my left hand and lying on my back, I got it all down, big and small.
The main problem of course was that my skin was ridiculously sensitive and painful. To ease this, I was covered in paraffin, from toes to neck. The paraffin is all over the inside of the synthetic hospital pyjamas. It soaks, somewhat, into the rasping synthetic sheets of the bed and synthetic pillowcase. The bed blankets are heavy and synthetic, and they too don’t mix well with paraffin. Everything is saturated with paraffin. None of it dries out at all or becomes any less slippery or welcoming. Under the paraffin, concreted to my skin are thousands of small golden crystals of solidified rash pus (see image for May 13). They are very hard and scratchy, and are difficult to dissolve in the shower. It is like having large grains of sand in the paraffin between me and the bed. The air conditioner is constantly on, blowing cool air onto me, and making the sheets cold and dead to the touch.
The skin pain is complimented by lots of things about my arms and hands that make it hard to do anything or be comfortable, apart from lying flat on my back. I have two cuts with stitches in them on my left forearm from the skin biopsies today. One of them has lost its dressing. I have a hospital name tag on my left wrist that is too loose. It will sound trivial, but it was extremely annoying. Hundreds of times, often using my mouth, I move it up and try to wedge it around my wrist, trying to keep it away from my forearm. The back of my left palm is bruised and sore from having a line in it for 3 days. The back of my right palm has been used about 10 times to draw blood. My right elbow crease area has the canula line in it, the tube going up into my vein above my elbow, making it uncomfortable to bend the arm. Two plastic dongle connectors hang from the line right to the exact middle of my ultra-sensitive forearm (see the image below). I need to find ways to position my arm so the plastic connectors are not resting on my forearm. Each time I move the arm, I try to avoid letting them touch me. The line hurts a little when the connectors dangle backwards out into the air, which happen if I raise the arm much (I am lying on my back, so raising the arm is a frequent need). The plastic bandage holding the line to my arm is half off, due to the paraffin, which makes it swing around more than it should.
There are about 60 blisters on my palms, between my fingers, and on the tops/bottoms of my fingers. Some are large, all are pressure sensitive. Closing my hands is awkward / sensitive due to this. Picking things up or, much harder, supporting my weight as I get up hurts the hands, for the same reason. I find it hard to bend down, let alone to reach the ground (skin pain) to pick anything up.
Lying on my back is fine, if I keep my arms angled out and clear of anything that could touch them or my underarms. But I have been lying down so much this past week, my back is very tired. Lying on my side is very uncomfortable due to arms, underarms, dongles, skin etc. I do it from time to time to relieve the back. I find a way to have both arms sticking out from my body, bent at the elbows, nothing touching anything, and I more-or-less hold the position and hope to drift off a little.
One thing I had been saving up and looking forward to as a midnight snack was a bowl of cereal with cold milk. After a couple of hours of lying in the bed, I decided it was time. I got up, feeling cold. I prepared the cereal and went to take the first delicious mouthful. Unbelievably, the milk had gone sour! I had to get warm again and the only place was the bed. But in the few minutes of preparing the cereal, I had left the sheets folded down and the air conditioner had blown on them. They were cold and saturated with paraffin. I forced myself back into the bed, pulled a blanket over myself, and tried to get warm.
At some point, I thought to myself “This is a survival course”. That everything that was going wrong, finishing with finding the milk sour and having to force myself into the wet sheets despite being freezing, was all part of a deliberate plan to break me. All part of a set of challenges I was being thrown and which I had to deal with. I mailed Russell, telling him I expected Derren Brown to walk into the room at any moment with a TV crew. The “survival course” perspective helped, and I began to smile (a little, inwardly). There was no way I was going to let this get the better of me. From then on, things have gotten better.
It was a horrible night. Probably the most uncomfortable of my life.
Friday May 17
My feet are now greatly swollen. Getting up from the bed is beginning to be painful. Negative test results come in on HIV and syphilis. During the day additional tests begin to come in.
Yesterday they weighed me: 81.6kg! I’ve somehow added 3.5kg in under 4 days. WTF?
Ana is worried I may have leukemia. It turns out she hasn’t slept for 3 days. The EMC guys have sent some worrying links, e.g., to things like Stevens Johnson disease. She brings me a ton of bedding: pillow cases, sheet, comforter cover, and a bunch of soft warm travel towels. That will make a huge difference.
Saturday May 18
It’s hard to see that I used to have ankles. My lower legs have turned dark purple with the rash and are very swollen. Large ugly blisters have appeared on inside of my heels, around my ankles, and on my Archilles tendons, on both feet The right leg is worse, but it’s a close thing. The upper part of my body is looking quite a bit better – compare the earlier photo of my underarm.
Dr Sterling drops in. She thinks, given the negative infectious disease results, it’s time to try some steroid cream on just my stomach & chest. She examines my “tree stump” legs and takes a photo with her phone.
Dr Moore drops by too. She looks at my legs and tells me that due to some cellular protein level being low, water that would otherwise be bound and inside my cells is leaving them via osmosis and is basically sloshing around (I am paraphrasing) inside my system, causing the inflammation of my feet. She tells me I have to eat well, with as much protein as possible. She suggests protein shakes. I tell Derek, who lives on SPIRU-TEIN, and he brings me a can.
Ana comes by and is still terribly worried I may have leukemia. I call to get a doctor in some that she can hear why they don’t think so. Mok, who works with Dr Moore, comes by to help. He says he’ll ask the lab doing the biopsy to examine the white blood cells (my count is high) for any abnormal shape that might indicate leukemia. Mok comes by later to lance some of the blisters on my heels to collect their liquid for analysis.
That night I have a fever of 39C. But they’d given me paracetamol and codeine which keeps it in check. The codeine makes me feel totally weird. I am aware of the distant sensory roar from my skin as I lie with my arms touching the sheets. I can’t think clearly about anything. I see faces transform into mischievous devil caricatures, with goatees and horns. At about 3am I manage to collect myself and put the steroid cream onto my chest. The codeine weirdness goes on all night, until daylight. In the morning I find the canula in the bed next to me, the tube has been pulled out of my vein. I’ll not be trying the codeine again.
Sunday May 19
Sofia and Lucas visit with Ana. We eat Burger King that they bring and watch a movie. They look a bit shocked to see my upper body. I keep my pants on so as not to show my legs, which are much scarier at this point.
Getting into a standing position is now very painful. Pain shoots down my legs as the “sloshing” intra-cellular water pushes down through my legs. Then when my feet touch the ground, some of the water is forced back up my legs and pains shoot up to my inner thigh. Once I manage to stand, I have to take small shuffling painful steps for maybe a minute before things settle down and I can walk almost normally. If this keeps getting worse, getting out of bed is going to be a real problem.
I have very small new blisters on fingers. What’s going on, more blisters?
Monday May 20
The creep of additional rash redness on feet has continued overnight, and the rash is now between toes. Walking in flip flops affected. I realize I should have been putting the steroid cream onto my entire foot, not just the parts that visually (only) have the rash.
Ana and Derek bring delicious Indian food from Cambridge for lunch.
More infectious disease test results are coming in, all negative.
Tuesday May 21
Althea and Edward Parker visit. Rash creep on hands and feet has stopped. Liver structural damage sonogram test is negative. A stool enterovirus test comes back negative, ruling out Hand, Foot and Mouth Disease. I am declared officially non-infective, and get to go downstairs to coffee shop!
Derek drops by at night and we talk virus discovery and a presentation I’m due to give in Rotterdam next Monday. I’ve done no Fluidinfo work or anything else for a couple of weeks.
I am peeling pretty much everywhere. Each morning and evening I wash myself and apply steroid cream. In between I am constantly putting on a skin itchiness/excema moisturizer.
Wednesday May 22
I thought I’d be out today because the key liver test is now showing an improved result. But they want to keep me one more day to do another blood test for liver functionality. I’m tempted to let them take blood in the morning and then check out. If the result is bad, I can come back in.
At Costa coffee downstairs I begin to pull together the images (mine, Derek’s, Ana’s) from the weirdness of the last 9 days. I do some work on putting together the text for this blog post. Costa have a fast and free wifi network which you can use for 3 hours.
My legs are peeling so much. There are many large flakes of skin in the bed each time I get in or out. The peeling skin on my heels and back of my Archilles tendon is very thick. You wouldn’t think that skin could peel, but it can. The skin on your balls (if you have any) can peel too, I can confirm. It feels like I can rub cream into my feet forever and they still could use more. From shoulders down to feet it looks like I have a mild sunburn, skin either peeling or very dry.
Thursday May 23
I’m supposed to have a last blood test today, but they’ve not come as they usually do to take the sample. I’m totally packed up and ready to leave. It’s been 10 days.
Now I’ve had word, I’ll be out soon. They’re preparing the various ointments I’ll need at home. Blood just got taken again and the results will be back in about an hour. Next Wednesday I’ll come back in for another blood test.
Conclusions
There’s no strong conclusion as to what caused my rash. The doctors think the most likely explanation is that it was triggered by a virus, but they don’t know what. It could have been medication, but apart from some cough syrup and lozenges, I wasn’t on any. They say it’s not an allergic reaction, and that it’s not due to the diet change. If they’re right, I’m lucky, as it’s unlikely to re-occur.
The Addenbrookes people have been fantastic. Perhaps 100 people have looked after me over the last ten days. Many doctors, nurses, blood takers, food bringers, cleaners, wheelchair pushers, x-ray and sonograph operators, admin staff. They’re from all over: Poland, Hungary, Latvia, Zimbabwe, South Africa, Australia, Ghana, UK, India, Pakistan, Philipines, China. Everyone has been great. The nurses work 12 hour shifts. In a few hours I’ll be gone from here and someone else will be in this room, being taken care of just as expertly as I was, and the good people working here just keep going and going and going, making each patient feel special and cared for, for thousands of patients a year in this ward alone. I’ve had an unpleasant last 10 days, but compared to what some people go through on a much longer term basis, it has been nothing. I’m lucky, I’ll walk out in almost full health and soon be fully recovered.
And thanks so much to Ana and Derek, for coming by every day and taking so much care of me. That made a huge difference.
Daylight robbery: Barclays skims €170 off a 5K EUR -> GBP transfer
13:23 February 5th, 2013 by terry. Posted under companies, me. | 4 Comments »
Last month (on Jan 18, 2013) someone I’m doing some work for initiated a transfer of €5,000 into my UK bank account. According to xe.com the mid-market rate that day was 1.1937940679 euros per pound.
So you might innocently expect to receive about 5,000 / 1.1937940679 = £4,188 minus any transfer fees.
The transfer went through an intermediate bank, who charged €17. Barclays charged “our commission” of a mere £6.
But the amount that arrived in my bank was not roughly £4,188 – £20 = £4,168 as you might hope.
The amount that arrived was £4017.74.
The friendly banks decided that the appropriate exchange rate for me that day was 1.23840, which is a full 4.5% higher than the mid-market 1.19379 rate. That’s £143 (€170).
Sure, I know there’s a buy/ask spread in currency and the mid-market rate isn’t what you’d get in any transaction. But taking £143 from your own customer just because you can is pretty fucking nasty. And so, via today’s arbitrary setting of the greed parameter in a bank computer, the voracious banking industry gobbles up just a little bit more of the money made by regular people. People who actually worked to earn that money.
It’s no wonder people hate their banks and that the financial system in general is so despised.